hclust2.py做聚类热图-基于MetaPhlAn结果或类似结构数据
MetaPhlAn怎么安装使用看这里: 202310-宏基组学物种分析工具-MetaPhlAn4安装和使用方法-Anaconda3- centos9 stream-CSDN博客
数据格式
MetaPhlAn运行完成后合并物种丰度结果,相同结构的矩阵也可以画聚类热图
head merged_abundance_class.txt
#直接出来的结果没有对齐使用excel给大家看直观一点分析作图
###就这么简单
source activate metaphlan4
hclust2.py -i merged_metaphlan_tables_class.txt -o merged_class.png怎么样? 很简单吧,其实作图就是这么简单,但实际上这个脚本还有很多其他参数,这些参数可以帮助大家对图片色彩,标签字体大小,聚类情况等做详细调整。大家在作图过程中需要根据自己情况进行反复调整。
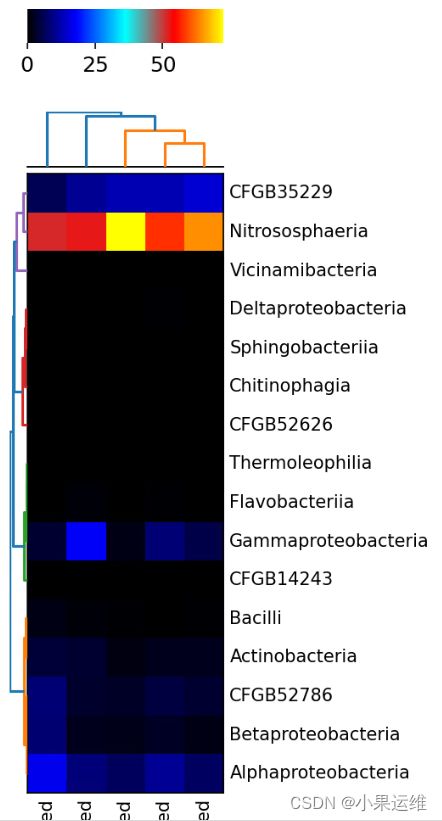
出图结果
下面就是出图结果,其他的自己看吧,
全参数帮助信息
###hclust2.py完整帮助文件:
hclust2.py -h
usage: hclust2.py [-h] [-i [INPUT_FILE]] [-o [OUTPUT_FILE]]
[--legend_file [LEGEND_FILE]] [-t INPUT_TYPE] [--sep SEP]
[--out_table OUT_TABLE] [--fname_row FNAME_ROW]
[--sname_row SNAME_ROW] [--metadata_rows METADATA_ROWS]
[--skip_rows SKIP_ROWS] [--sperc SPERC] [--fperc FPERC]
[--stop STOP] [--ftop FTOP] [--def_na DEF_NA]
[--f_dist_f F_DIST_F] [--s_dist_f S_DIST_F]
[--load_dist_matrix_f LOAD_DIST_MATRIX_F]
[--load_dist_matrix_s LOAD_DIST_MATRIX_S]
[--load_pickled_dist_matrix_f LOAD_PICKLED_DIST_MATRIX_F]
[--load_pickled_dist_matrix_s LOAD_PICKLED_DIST_MATRIX_S]
[--save_pickled_dist_matrix_f SAVE_PICKLED_DIST_MATRIX_F]
[--save_pickled_dist_matrix_s SAVE_PICKLED_DIST_MATRIX_S]
[--no_fclustering] [--no_plot_fclustering]
[--no_sclustering] [--no_plot_sclustering]
[--flinkage FLINKAGE] [--slinkage SLINKAGE] [--dpi DPI] [-l]
[--title TITLE] [--title_fontsize TITLE_FONTSIZE] [-s]
[--no_slabels] [--minv MINV] [--maxv MAXV] [--no_flabels]
[--max_slabel_len MAX_SLABEL_LEN]
[--max_flabel_len MAX_FLABEL_LEN]
[--flabel_size FLABEL_SIZE] [--slabel_size SLABEL_SIZE]
[--fdend_width FDEND_WIDTH] [--sdend_height SDEND_HEIGHT]
[--metadata_height METADATA_HEIGHT]
[--metadata_separation METADATA_SEPARATION]
[--colorbar_font_size COLORBAR_FONT_SIZE]
[--image_size IMAGE_SIZE]
[--cell_aspect_ratio CELL_ASPECT_RATIO]
[-c {Blues,BrBG,BuGn,BuPu,GnBu,Greens,Greys,OrRd,Oranges,PRGn,PiYG,PuBu,PuBuGn,PuOr,PuRd,Purples,RdBu,RdGy,RdPu,RdYlBu,RdYlGn,Reds,Spectral,YlGn,YlGnBu,YlOrBr,YlOrRd,afmhot,autumn,binary,bone,brg,bwr,cool,copper,flag,gist_earth,gist_gray,gist_heat,gist_ncar,gist_rainbow,gist_stern,gist_yarg,gnuplot,gnuplot2,gray,hot,hsv,jet,ocean,pink,prism,rainbow,seismic,spectral,spring,summer,terrain,winter,bbcyr,bbcry,bcry}]
[--bottom_c BOTTOM_C] [--top_c TOP_C] [--nan_c NAN_C]
TBA
optional arguments:
-h, --help show this help message and exit
-i [INPUT_FILE], --inp [INPUT_FILE], --in [INPUT_FILE]
The input matrix
-o [OUTPUT_FILE], --out [OUTPUT_FILE]
The output image file [image on screen of not
specified]
--legend_file [LEGEND_FILE]
The output file for the legend of the provided
metadata
-t INPUT_TYPE, --input_type INPUT_TYPE
The input type can be a data matrix or distance matrix
[default data_matrix]
Input data matrix parameters:
--sep SEP
--out_table OUT_TABLE
Write processed data matrix to file
--fname_row FNAME_ROW
row number containing the names of the features
[default 0, specify -1 if no names are present in the
matrix
--sname_row SNAME_ROW
column number containing the names of the samples
[default 0, specify -1 if no names are present in the
matrix
--metadata_rows METADATA_ROWS
Row numbers to use as metadata[default None, meaning
no metadata
--skip_rows SKIP_ROWS
Row numbers to skip (0-indexed, comma separated) from
the input file[default None, meaning no rows skipped
--sperc SPERC Percentile of sample value distribution for sample
selection
--fperc FPERC Percentile of feature value distribution for sample
selection
--stop STOP Number of top samples to select (ordering based on
percentile specified by --sperc)
--ftop FTOP Number of top features to select (ordering based on
percentile specified by --fperc)
--def_na DEF_NA Set the default value for missing values [default None
which means no replacement]
Distance parameters:
--f_dist_f F_DIST_F Distance function for features [default correlation]
--s_dist_f S_DIST_F Distance function for sample [default euclidean]
--load_dist_matrix_f LOAD_DIST_MATRIX_F
Load the distance matrix to be used for features
[default None].
--load_dist_matrix_s LOAD_DIST_MATRIX_S
Load the distance matrix to be used for samples
[default None].
--load_pickled_dist_matrix_f LOAD_PICKLED_DIST_MATRIX_F
Load the distance matrix to be used for features as
previously saved as pickle file using hclust2 itself
[default None].
--load_pickled_dist_matrix_s LOAD_PICKLED_DIST_MATRIX_S
Load the distance matrix to be used for samples as
previously saved as pickle file using hclust2 itself
[default None].
--save_pickled_dist_matrix_f SAVE_PICKLED_DIST_MATRIX_F
Save the distance matrix for features to file [default
None].
--save_pickled_dist_matrix_s SAVE_PICKLED_DIST_MATRIX_S
Save the distance matrix for samples to file [default
None].
Clustering parameters:
--no_fclustering avoid clustering features
--no_plot_fclustering
avoid plotting the feature dendrogram
--no_sclustering avoid clustering samples
--no_plot_sclustering
avoid plotting the sample dendrogram
--flinkage FLINKAGE Linkage method for feature clustering [default
average]
--slinkage SLINKAGE Linkage method for sample clustering [default average]
Heatmap options:
--dpi DPI Image resolution in dpi [default 150]
-l, --log_scale Log scale
--title TITLE Title of the plot
--title_fontsize TITLE_FONTSIZE
Font size of the title
-s, --sqrt_scale Square root scale
--no_slabels Do not show sample labels
--minv MINV Minimum value to display in the color map [default
None meaning automatic]
--maxv MAXV Maximum value to display in the color map [default
None meaning automatic]
--no_flabels Do not show feature labels
--max_slabel_len MAX_SLABEL_LEN
Max number of chars to report for sample labels
[default 15]
--max_flabel_len MAX_FLABEL_LEN
Max number of chars to report for feature labels
[default 15]
--flabel_size FLABEL_SIZE
Feature label font size [default 10]
--slabel_size SLABEL_SIZE
Sample label font size [default 10]
--fdend_width FDEND_WIDTH
Width of the feature dendrogram [default 1 meaning
100% of default heatmap width]
--sdend_height SDEND_HEIGHT
Height of the sample dendrogram [default 1 meaning
100% of default heatmap height]
--metadata_height METADATA_HEIGHT
Height of the metadata panel [default 0.05 meaning 5%
of default heatmap height]
--metadata_separation METADATA_SEPARATION
Distance between the metadata and data panels.
[default 0.001 meaning 0.1% of default heatmap height]
--colorbar_font_size COLORBAR_FONT_SIZE
Color bar label font size [default 12]
--image_size IMAGE_SIZE
Size of the largest between width and eight size for
the image in inches [default 8]
--cell_aspect_ratio CELL_ASPECT_RATIO
Aspect ratio between width and height for the cells of
the heatmap [default 1.0]
-c {Blues,BrBG,BuGn,BuPu,GnBu,Greens,Greys,OrRd,Oranges,PRGn,PiYG,PuBu,PuBuGn,PuOr,PuRd,Purples,RdBu,RdGy,RdPu,RdYlBu,RdYlGn,Reds,Spectral,YlGn,YlGnBu,YlOrBr,YlOrRd,afmhot,autumn,binary,bone,brg,bwr,cool,copper,flag,gist_earth,gist_gray,gist_heat,gist_ncar,gist_rainbow,gist_stern,gist_yarg,gnuplot,gnuplot2,gray,hot,hsv,jet,ocean,pink,prism,rainbow,seismic,spectral,spring,summer,terrain,winter,bbcyr,bbcry,bcry}, --colormap {Blues,BrBG,BuGn,BuPu,GnBu,Greens,Greys,OrRd,Oranges,PRGn,PiYG,PuBu,PuBuGn,PuOr,PuRd,Purples,RdBu,RdGy,RdPu,RdYlBu,RdYlGn,Reds,Spectral,YlGn,YlGnBu,YlOrBr,YlOrRd,afmhot,autumn,binary,bone,brg,bwr,cool,copper,flag,gist_earth,gist_gray,gist_heat,gist_ncar,gist_rainbow,gist_stern,gist_yarg,gnuplot,gnuplot2,gray,hot,hsv,jet,ocean,pink,prism,rainbow,seismic,spectral,spring,summer,terrain,winter,bbcyr,bbcry,bcry}
--bottom_c BOTTOM_C Color to use for cells below the minimum value of the
scale [default None meaning bottom color of the scale]
--top_c TOP_C Color to use for cells below the maximum value of the
scale [default None meaning bottom color of the scale]
--nan_c NAN_C Color to use for nan cells [default None]
常用调整参数
--stop 当样品数太多,也就是列数太多,可选择自己希望显示丰度最高的前多个个样品
--ftop 当物种数太多,也就是行数太多,可以选择自己希望显示的丰度最高的多少个物种
--dpi 这个很重要哦,当行列数比较多时需要自己调整整个输出图的画幅以适应实际需求
--flabel_size 这里就是行头物种名标签在图片中的大小,
--slabel_size 同样这里是表头中样品名标签在图片中的大小。
--cell_aspect_ratio 这个就是上面显示的每个方格的长和宽的比例,默认为1:1也就是方块,0.5也即是横向的长方形了,2就成了纵向的长方形了,这个可以将图调整成合适的比例显示。
--no_fclustering --no_sclustering 这个很重要,有时候不想查看样品之间的聚类情况,使用--no_clustering后图中的样品会按顺序出现在图中,同样--no_fclustering则是不进行物种的聚类。
-c 这个参数是指图片显示色彩的体系,默认为红-蓝色系,可以指定其他的,自己试着选就行了。
其他的大家参考全参数帮助文件吧,大家对照着调整即可;个人一般会反复调整到合适的比例和色彩