PSP - 蛋白质复合物结构预测 模版配对(Template Pair) 逻辑的特征分析
欢迎关注我的CSDN:https://spike.blog.csdn.net/
本文地址:https://spike.blog.csdn.net/article/details/134328447
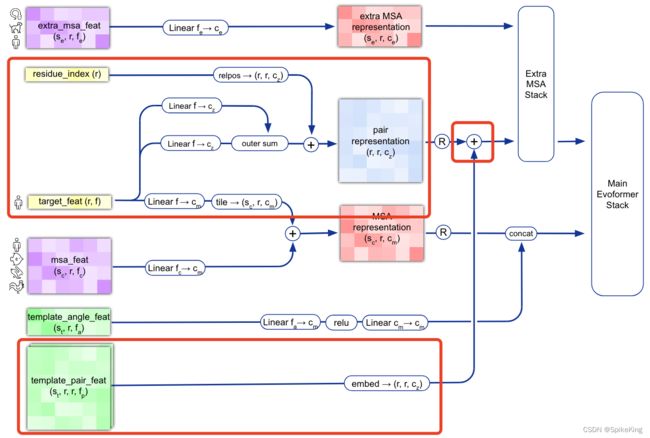
在 蛋白质复合物结构预测 的过程中,模版 (Template) 起到重要作用,提供预测结果的关于三维结构的先验信息,在多链的情况,需要进行模版配对,即 Template Pair,核心函数是 template_pair_embedder,结合 AlphaFold2 的论文,分析具体输入与输出的特征。
核心逻辑 template_pair_embedder(),输入和输出特征维度:
[CL] TemplateEmbedderMultimer - template_dgram: torch.Size([1, 1102, 1102, 39])
[CL] TemplateEmbedderMultimer - z: torch.Size([1102, 1102, 128])
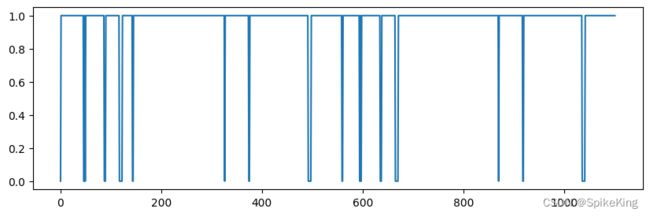
[CL] TemplateEmbedderMultimer - pseudo_beta_mask: torch.Size([1, 1102])
[CL] TemplateEmbedderMultimer - backbone_mask: torch.Size([1, 1102])
[CL] TemplateEmbedderMultimer - multichain_mask_2d: torch.Size([1102, 1102])
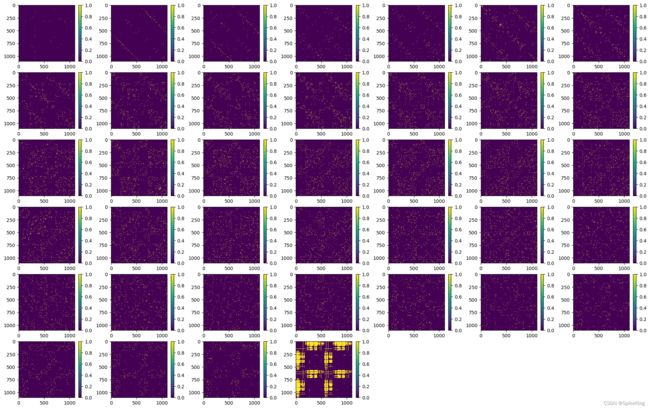
[CL] TemplateEmbedderMultimer - unit_vector: torch.Size([1, 1102, 1102])
[CL] TemplateEmbedderMultimer - pair_act: torch.Size([1, 1102, 1102, 64])
函数:
# openfold/model/embedders.py
pair_act = self.template_pair_embedder(
template_dgram,
aatype_one_hot,
z,
pseudo_beta_mask,
backbone_mask,
multichain_mask_2d,
unit_vector,
)
t = torch.sum(t, dim=-4) / n_templ
t = torch.nn.functional.relu(t)
t = self.linear_t(t) # 从 c_t 维度 转换 成 c_z 维度,更新 z
template_embeds["template_pair_embedding"] = t
# openfold/model/model.py
template_embeds = self.template_embedder(
template_feats,
z,
pair_mask.to(dtype=z.dtype),
no_batch_dims,
chunk_size=self.globals.chunk_size,
multichain_mask_2d=multichain_mask_2d,
use_fa=self.globals.use_fa,
)
z = z + template_embeds["template_pair_embedding"] # line 13 in Alg.2
逻辑图:
1. template_dgram 特征
template_dgram特征计算,不同点距离其他点的远近划分,共划分 39 个bin,Template 的 no_bin 是 1.25 计算 1 个值,即 (50.75 - 3.25) / 38 = 1.25,即:
template_dgram = dgram_from_positions(
template_positions,
inf=self.config.inf,
**self.config.distogram,
)
def dgram_from_positions(
pos: torch.Tensor,
min_bin: float = 3.25,
max_bin: float = 50.75,
no_bins: float = 39,
inf: float = 1e8,
):
dgram = torch.sum(
(pos[..., None, :] - pos[..., None, :, :]) ** 2, dim=-1, keepdim=True
)
lower = torch.linspace(min_bin, max_bin, no_bins, device=pos.device) ** 2
upper = torch.cat([lower[1:], lower.new_tensor([inf])], dim=-1)
dgram = ((dgram > lower) * (dgram < upper)).type(dgram.dtype)
return dgram
Template 的 no_bin 是 1.25 计算 1 个值,即 (50.75 - 3.25) / 38 = 1.25,长度是 39。
其中,template_positions 特征,如下:
- 输入
template_pseudo_beta与template_pseudo_beta_mask - 输出坐标
template_positions,即template_pseudo_beta的值
template_positions, pseudo_beta_mask = (
single_template_feats["template_pseudo_beta"],
single_template_feats["template_pseudo_beta_mask"],
)
其中pseudo_beta 特征的处理,来源于 openfold/data/data_transforms_multimer.py 即:
- 输入特征:
template_aatype、template_all_atom_positions、template_all_atom_mask - 原理是:选择 CA 或 CB 原子的坐标与 Mask
源码即调用关系如下:
# 输入特征,模型预测结构
# run_pretrained_openfold.py
processed_feature_dict, _ = feature_processor.process_features(
feature_dict, is_multimer, mode="predict"
)
output_dict = predict_structure_single_dev(
args,
model_name,
current_model,
fasta_path,
processed_feature_dict,
config,
)
# openfold/data/feature_pipeline.py
processed_feature, label = np_example_to_features_multimer(
np_example=raw_features,
config=self.config,
mode=mode,
)
# openfold/data/feature_pipeline.py
features, label = input_pipeline_multimer.process_tensors_from_config(
tensor_dict,
cfg.common,
cfg[mode],
cfg.data_module,
)
# openfold/data/input_pipeline_multimer.py
nonensembled = nonensembled_transform_fns(
common_cfg,
mode_cfg,
)
tensors = compose(nonensembled)(tensors)
# openfold/data/input_pipeline_multimer.py
operators.extend(
[
data_transforms_multimer.make_atom14_positions,
data_transforms_multimer.atom37_to_frames,
data_transforms_multimer.atom37_to_torsion_angles(""),
data_transforms_multimer.make_pseudo_beta(""),
data_transforms_multimer.get_backbone_frames,
data_transforms_multimer.get_chi_angles,
]
)
# openfold/data/data_transforms_multimer.py
def make_pseudo_beta(protein, prefix=""):
"""Create pseudo-beta (alpha for glycine) position and mask."""
assert prefix in ["", "template_"]
(
protein[prefix + "pseudo_beta"],
protein[prefix + "pseudo_beta_mask"],
) = pseudo_beta_fn(
protein["template_aatype" if prefix else "aatype"],
protein[prefix + "all_atom_positions"],
protein["template_all_atom_mask" if prefix else "all_atom_mask"],
)
return protein
# openfold/data/data_transforms_multimer.py
def pseudo_beta_fn(aatype, all_atom_positions, all_atom_mask):
"""Create pseudo beta features."""
if aatype.shape[0] > 0:
is_gly = torch.eq(aatype, rc.restype_order["G"])
ca_idx = rc.atom_order["CA"]
cb_idx = rc.atom_order["CB"]
pseudo_beta = torch.where(
torch.tile(is_gly[..., None], [1] * len(is_gly.shape) + [3]),
all_atom_positions[..., ca_idx, :],
all_atom_positions[..., cb_idx, :],
)
else:
pseudo_beta = all_atom_positions.new_zeros(*aatype.shape, 3)
if all_atom_mask is not None:
if aatype.shape[0] > 0:
pseudo_beta_mask = torch.where(
is_gly, all_atom_mask[..., ca_idx], all_atom_mask[..., cb_idx]
)
else:
pseudo_beta_mask = torch.zeros_like(aatype).float()
return pseudo_beta, pseudo_beta_mask
else:
return pseudo_beta
template_pseudo_beta_mask: Mask indicating if the beta carbon (alpha carbon for glycine) atom has coordinates for the template at this residue.
template_dgram 特征 [1, 1102, 1102, 39],即:
2. z 特征
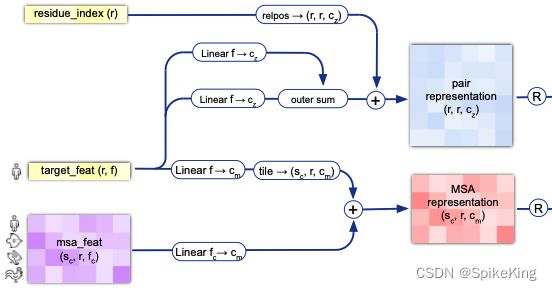
z 特征作为输入,直接传入,来源于 protein["target_feat"],来源于protein[]"between_segment_residues"], 即:
- 日志
target_feat:torch.Size([1102, 21]),不包括 “-”,只包括21=20+1个氨基酸,包括X - 将
[1102, 21]经过线性层,转换成c_z维度,即128维。 - 再通过
outer sum操作 转换成 LxLxC 维度,其实 z 就是 Pair Representation,即[1102, 1102, 128]维度。
# openfold/model/embedders.py
def forward(
self,
batch,
z,
padding_mask_2d,
templ_dim,
chunk_size,
multichain_mask_2d,
use_fa=False,
):
# openfold/model/model.py
template_embeds = self.template_embedder(
template_feats,
z,
pair_mask.to(dtype=z.dtype),
no_batch_dims,
chunk_size=self.globals.chunk_size,
multichain_mask_2d=multichain_mask_2d,
use_fa=self.globals.use_fa,
)
# openfold/model/model.py
m, z = self.input_embedder(feats)
# openfold/model/embedders.py#InputEmbedderMultimer
def forward(self, batch) -> Tuple[torch.Tensor, torch.Tensor]:
"""
# ...
Returns:
msa_emb:
[*, N_clust, N_res, C_m] MSA embedding
pair_emb:
[*, N_res, N_res, C_z] pair embedding
"""
tf = batch["target_feat"]
msa = batch["msa_feat"]
# [*, N_res, c_z]
tf_emb_i = self.linear_tf_z_i(tf)
tf_emb_j = self.linear_tf_z_j(tf)
# [*, N_res, N_res, c_z]
pair_emb = tf_emb_i[..., None, :] + tf_emb_j[..., None, :, :]
pair_emb = pair_emb + self.relpos(batch) # 计算相对位置
# [*, N_clust, N_res, c_m]
n_clust = msa.shape[-3]
tf_m = (
self.linear_tf_m(tf)
.unsqueeze(-3)
.expand(((-1,) * len(tf.shape[:-2]) + (n_clust, -1, -1)))
)
msa_emb = self.linear_msa_m(msa) + tf_m
return msa_emb, pair_emb
# openfold/data/data_transforms_multimer.py
def create_target_feat(batch):
"""Create the target features"""
batch["target_feat"] = torch.nn.functional.one_hot(batch["aatype"], 21).to(
torch.float32
)
return batch
# openfold/data/input_pipeline_multimer.py
operators.extend(
[
data_transforms_multimer.cast_to_64bit_ints,
# todo: randomly_replace_msa_with_unknown may need to be confirmed and tried in training.
# data_transforms_multimer.randomly_replace_msa_with_unknown(0.0),
data_transforms_multimer.make_msa_profile,
data_transforms_multimer.create_target_feat,
data_transforms_multimer.make_atom14_masks,
]
)
其中的 21 个氨基酸,即:
ID_TO_HHBLITS_AA = {
0: "A",
1: "C", # Also U.
2: "D", # Also B.
3: "E", # Also Z.
4: "F",
5: "G",
6: "H",
7: "I",
8: "K",
9: "L",
10: "M",
11: "N",
12: "P",
13: "Q",
14: "R",
15: "S",
16: "T",
17: "V",
18: "W",
19: "Y",
20: "X", # Includes J and O.
21: "-",
}
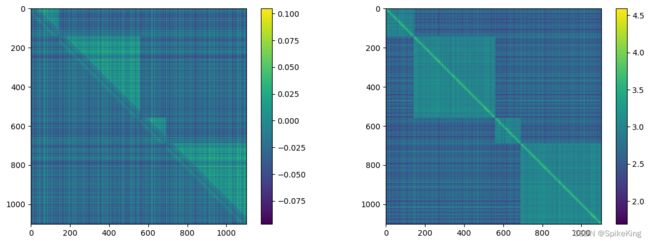
z 特征 (mean 和 max),[1102, 1102, 128],即:
3. pseudo_beta_mask 与 backbone_mask 特征
pseudo_beta_mask 特征 参考 template_dgram 的 template_pseudo_beta 部分的源码:
- 关注 CA 与 CB 的 Mask 信息
# openfold/data/data_transforms_multimer.py
def make_pseudo_beta(protein, prefix=""):
"""Create pseudo-beta (alpha for glycine) position and mask."""
assert prefix in ["", "template_"]
(
protein[prefix + "pseudo_beta"],
protein[prefix + "pseudo_beta_mask"],
) = pseudo_beta_fn(
protein["template_aatype" if prefix else "aatype"],
protein[prefix + "all_atom_positions"],
protein["template_all_atom_mask" if prefix else "all_atom_mask"],
)
return protein
# openfold/data/data_transforms_multimer.py
def pseudo_beta_fn(aatype, all_atom_positions, all_atom_mask):
"""Create pseudo beta features."""
if aatype.shape[0] > 0:
is_gly = torch.eq(aatype, rc.restype_order["G"])
ca_idx = rc.atom_order["CA"]
cb_idx = rc.atom_order["CB"]
pseudo_beta = torch.where(
torch.tile(is_gly[..., None], [1] * len(is_gly.shape) + [3]),
all_atom_positions[..., ca_idx, :],
all_atom_positions[..., cb_idx, :],
)
else:
pseudo_beta = all_atom_positions.new_zeros(*aatype.shape, 3)
if all_atom_mask is not None:
if aatype.shape[0] > 0:
pseudo_beta_mask = torch.where(
is_gly, all_atom_mask[..., ca_idx], all_atom_mask[..., cb_idx]
)
else:
pseudo_beta_mask = torch.zeros_like(aatype).float()
return pseudo_beta, pseudo_beta_mask
else:
return pseudo_beta
其中,openfold/data/msa_pairing.py#merge_chain_features(), 合并(merge)多链特征,输出 Template Feature,即:
[CL] template features, template_aatype : (4, 1102)
[CL] template features, template_all_atom_positions : (4, 1102, 37, 3)
[CL] template features, template_all_atom_mask : (4, 1102, 37)
注意 template_all_atom_mask,即 37 个原子的 mask 信息。
其中,37 个原子(Atom):
{'N': 0, 'CA': 1, 'C': 2, 'CB': 3, 'O': 4, 'CG': 5, 'CG1': 6, 'CG2': 7, 'OG': 8, 'OG1': 9, 'SG': 10,
'CD': 11, 'CD1': 12, 'CD2': 13, 'ND1': 14, 'ND2': 15, 'OD1': 16, 'OD2': 17, 'SD': 18, 'CE': 19, 'CE1': 20,
'CE2': 21, 'CE3': 22, 'NE': 23, 'NE1': 24, 'NE2': 25, 'OE1': 26, 'OE2': 27, 'CH2': 28, 'NH1': 29, 'NH2': 30,
'OH': 31, 'CZ': 32, 'CZ2': 33, 'CZ3': 34, 'NZ': 35, 'OXT': 36}
backbone_mask 特征,只关注 N、CA、C 三类原子的 Mask 信息,参考:
- 一般情况下都与
pseudo_beta_mask相同,因为要么存在残基,要么不存在残基。
# openfold/utils/all_atom_multimer.py
def make_backbone_affine(
positions: geometry.Vec3Array,
mask: torch.Tensor,
aatype: torch.Tensor,
) -> Tuple[geometry.Rigid3Array, torch.Tensor]:
a = rc.atom_order["N"]
b = rc.atom_order["CA"]
c = rc.atom_order["C"]
rigid_mask = mask[..., a] * mask[..., b] * mask[..., c]
rigid = make_transform_from_reference(
a_xyz=positions[..., a],
b_xyz=positions[..., b],
c_xyz=positions[..., c],
)
return rigid, rigid_mask
pseudo_beta_mask 与 backbone_mask 相同,[1, 1102],即:
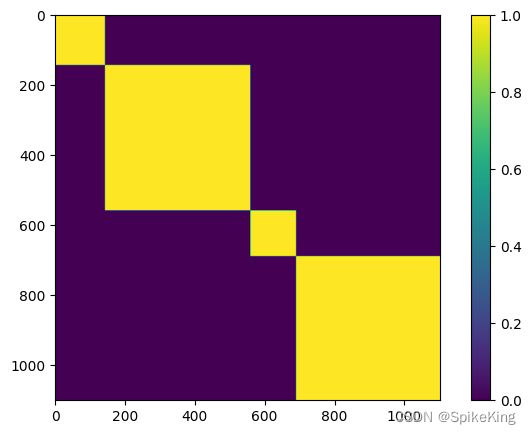
4. multichain_mask_2d 特征
非常简单,就是链内 Mask,源码:
# openfold/model/model.py
multichain_mask_2d = (
asym_id[..., None] == asym_id[..., None, :]
) # [N_res, N_res]
multichain_mask_2d,[1102, 1102],即:
5. unit_vector 特征
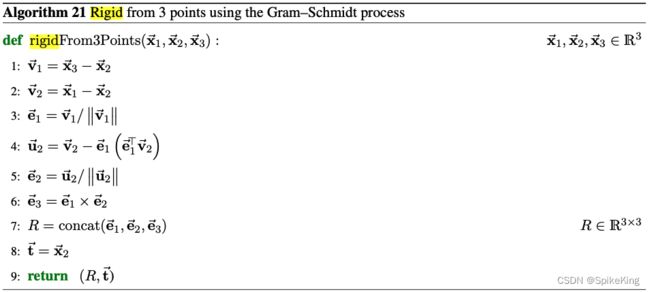
unit_vector 是 Rot3Array 对象,与角度相关的单位向量,源码:
# openfold/model/embedders.py
rigid, backbone_mask = all_atom_multimer.make_backbone_affine(
atom_pos,
single_template_feats["template_all_atom_mask"],
single_template_feats["template_aatype"],
)
points = rigid.translation
rigid_vec = rigid[..., None].inverse().apply_to_point(points)
unit_vector = rigid_vec.normalized()
# openfold/utils/all_atom_multimer.py
def make_backbone_affine(
positions: geometry.Vec3Array,
mask: torch.Tensor,
aatype: torch.Tensor,
) -> Tuple[geometry.Rigid3Array, torch.Tensor]:
a = rc.atom_order["N"]
b = rc.atom_order["CA"]
c = rc.atom_order["C"]
rigid_mask = mask[..., a] * mask[..., b] * mask[..., c]
rigid = make_transform_from_reference(
a_xyz=positions[..., a],
b_xyz=positions[..., b],
c_xyz=positions[..., c],
)
return rigid, rigid_mask
# openfold/utils/all_atom_multimer.py
def make_transform_from_reference(
a_xyz: geometry.Vec3Array, b_xyz: geometry.Vec3Array, c_xyz: geometry.Vec3Array
) -> geometry.Rigid3Array:
"""Returns rotation and translation matrices to convert from reference.
Note that this method does not take care of symmetries. If you provide the
coordinates in the non-standard way, the A atom will end up in the negative
y-axis rather than in the positive y-axis. You need to take care of such
cases in your code.
Args:
a_xyz: A Vec3Array.
b_xyz: A Vec3Array.
c_xyz: A Vec3Array.
Returns:
A Rigid3Array which, when applied to coordinates in a canonicalized
reference frame, will give coordinates approximately equal
the original coordinates (in the global frame).
"""
rotation = geometry.Rot3Array.from_two_vectors(c_xyz - b_xyz, a_xyz - b_xyz)
return geometry.Rigid3Array(rotation, b_xyz)
# openfold/utils/geometry/rotation_matrix.py
@classmethod
def from_two_vectors(cls, e0: vector.Vec3Array, e1: vector.Vec3Array) -> Rot3Array:
"""Construct Rot3Array from two Vectors.
Rot3Array is constructed such that in the corresponding frame 'e0' lies on
the positive x-Axis and 'e1' lies in the xy plane with positive sign of y.
Args:
e0: Vector
e1: Vector
Returns:
Rot3Array
"""
# Normalize the unit vector for the x-axis, e0.
e0 = e0.normalized()
# make e1 perpendicular to e0.
c = e1.dot(e0)
e1 = (e1 - c * e0).normalized()
# Compute e2 as cross product of e0 and e1.
e2 = e0.cross(e1)
return cls(e0.x, e1.x, e2.x, e0.y, e1.y, e2.y, e0.z, e1.z, e2.z)
解释:每个残基的局部框架内所有残基的 α \alpha α 碳原子位移的单位向量。 这些局部框架的计算方式与目标结构相同。
The unit vector of the displacement of the alpha carbon atom of all residues within the local frame of each residue. These local frames are computed in the same way as for the target structure.
计算逻辑:
unit_vector 包括x、y、z等3个分量,[1, 1102, 1102, 3],即: