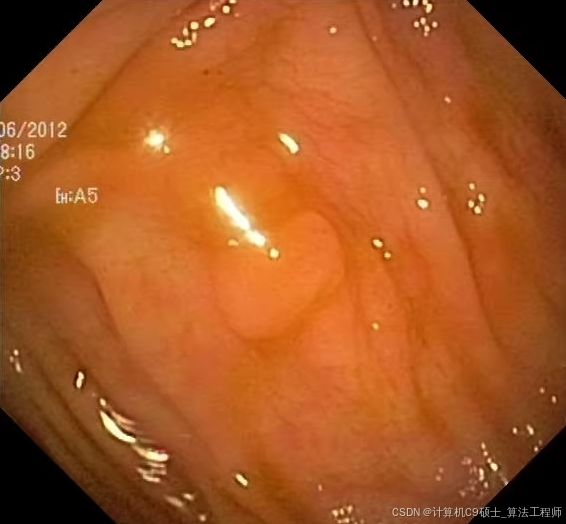
医学类 使用TransUNet、UNet、DeepLabV3+、HRNet、PSPNet 模型对息肉分割数据集进行训练、评估和可视化 EDD2020息肉数据集分割数据集
息肉数据集/息肉瘤分割项目解决(已处理好:
EDD2020数据集(Endoscopy Disease Detection and Segmentation Challenge)
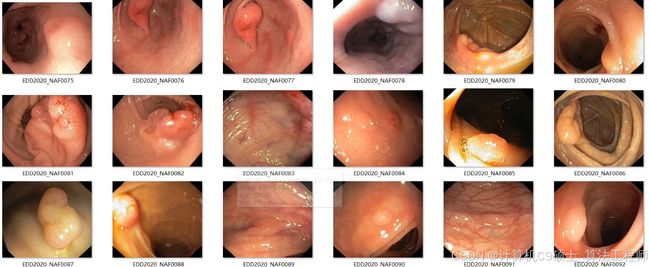
该息肉分割数据集主要包含人体生长的(肠胃)息肉
用于器官内部 息肉瘤分割,息肉目标检测,息肉定位任务
息肉分割是一个重要的医学影像分析任务,特别是在内窥镜检查中。EDD2020数据集是一个很好的起点。我们将使用几种流行的深度学习模型(如TransUNet、UNet、DeepLabV3+、HRNet、PSPNet)来进行息肉分割。以下是详细的步骤和代码示例。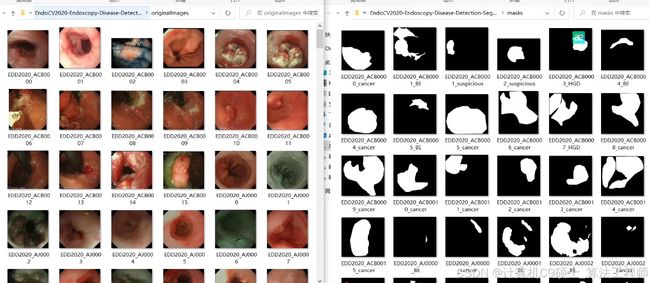
1. 环境准备
首先,确保你已经安装了必要的库和工具。你可以使用以下命令安装所需的库:
pip install torch torchvision
pip install numpy
pip install pandas
pip install matplotlib
pip install opencv-python
pip install segmentation-models-pytorch
pip install albumentations
2. 数据集准备
假设你的数据集目录结构如下:
polyp_dataset/
├── images/
│ ├── train/
│ ├── val/
│ └── test/
├── masks/
│ ├── train/
│ ├── val/
│ └── test/
└── polyp.yaml
每个图像文件和对应的掩码文件都以相同的文件名命名,例如 0001.png 和 0001.png。
3. 创建数据集配置文件
你已经有一个 polyp.yaml 文件,内容如下:
train: ../polyp_dataset/images/train
val: ../polyp_dataset/images/val
test: ../polyp_dataset/images/test
mask_train: ../polyp_dataset/masks/train
mask_val: ../polyp_dataset/masks/val
mask_test: ../polyp_dataset/masks/test
nc: 1
names: ['polyp']
4. 数据加载和预处理
使用PyTorch和Albumentations进行数据加载和预处理。
import os
import cv2
import numpy as np
import torch
from torch.utils.data import Dataset, DataLoader
import albumentations as A
from albumentations.pytorch import ToTensorV2
class PolypDataset(Dataset):
def __init__(self, image_dir, mask_dir, transform=None):
self.image_dir = image_dir
self.mask_dir = mask_dir
self.transform = transform
self.images = os.listdir(image_dir)
def __len__(self):
return len(self.images)
def __getitem__(self, index):
img_path = os.path.join(self.image_dir, self.images[index])
mask_path = os.path.join(self.mask_dir, self.images[index])
image = cv2.imread(img_path, cv2.IMREAD_COLOR)
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
mask = cv2.imread(mask_path, cv2.IMREAD_GRAYSCALE)
mask = mask / 255.0
if self.transform is not None:
augmented = self.transform(image=image, mask=mask)
image = augmented['image']
mask = augmented['mask']
return image, mask
# 数据增强
transform = A.Compose([
A.Resize(height=256, width=256),
A.Normalize(mean=(0.485, 0.456, 0.406), std=(0.229, 0.224, 0.225)),
ToTensorV2()
])
# 加载数据集
train_dataset = PolypDataset(
image_dir='polyp_dataset/images/train',
mask_dir='polyp_dataset/masks/train',
transform=transform
)
val_dataset = PolypDataset(
image_dir='polyp_dataset/images/val',
mask_dir='polyp_dataset/masks/val',
transform=transform
)
test_dataset = PolypDataset(
image_dir='polyp_dataset/images/test',
mask_dir='polyp_dataset/masks/test',
transform=transform
)
train_loader = DataLoader(train_dataset, batch_size=4, shuffle=True, num_workers=4)
val_loader = DataLoader(val_dataset, batch_size=4, shuffle=False, num_workers=4)
test_loader = DataLoader(test_dataset, batch_size=4, shuffle=False, num_workers=4)
5. 模型选择和训练
我们将使用几种流行的分割模型进行训练。以下是使用UNet和DeepLabV3+的示例。
UNet
import segmentation_models_pytorch as smp
# 定义模型
model = smp.Unet(
encoder_name="resnet34", # 选择预训练的编码器
encoder_weights="imagenet", # 使用ImageNet预训练权重
in_channels=3, # 输入通道数
classes=1 # 输出通道数
)
# 定义损失函数和优化器
criterion = torch.nn.BCEWithLogitsLoss()
optimizer = torch.optim.Adam(model.parameters(), lr=0.001)
# 训练函数
def train_model(model, criterion, optimizer, train_loader, val_loader, num_epochs=25):
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
model.to(device)
for epoch in range(num_epochs):
model.train()
running_loss = 0.0
for images, masks in train_loader:
images, masks = images.to(device), masks.to(device)
optimizer.zero_grad()
outputs = model(images)
loss = criterion(outputs, masks.unsqueeze(1))
loss.backward()
optimizer.step()
running_loss += loss.item() * images.size(0)
train_loss = running_loss / len(train_loader.dataset)
model.eval()
running_val_loss = 0.0
with torch.no_grad():
for images, masks in val_loader:
images, masks = images.to(device), masks.to(device)
outputs = model(images)
loss = criterion(outputs, masks.unsqueeze(1))
running_val_loss += loss.item() * images.size(0)
val_loss = running_val_loss / len(val_loader.dataset)
print(f"Epoch {epoch + 1}/{num_epochs}, Train Loss: {train_loss:.4f}, Val Loss: {val_loss:.4f}")
# 训练模型
train_model(model, criterion, optimizer, train_loader, val_loader, num_epochs=25)
DeepLabV3+
# 定义模型
model = smp.DeepLabV3Plus(
encoder_name="resnet34", # 选择预训练的编码器
encoder_weights="imagenet", # 使用ImageNet预训练权重
in_channels=3, # 输入通道数
classes=1 # 输出通道数
)
# 定义损失函数和优化器
criterion = torch.nn.BCEWithLogitsLoss()
optimizer = torch.optim.Adam(model.parameters(), lr=0.001)
# 训练模型
train_model(model, criterion, optimizer, train_loader, val_loader, num_epochs=25)
6. 评估模型
使用测试数据评估模型性能。
def evaluate_model(model, test_loader, criterion):
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
model.to(device)
model.eval()
running_loss = 0.0
with torch.no_grad():
for images, masks in test_loader:
images, masks = images.to(device), masks.to(device)
outputs = model(images)
loss = criterion(outputs, masks.unsqueeze(1))
running_loss += loss.item() * images.size(0)
test_loss = running_loss / len(test_loader.dataset)
print(f"Test Loss: {test_loss:.4f}")
# 评估模型
evaluate_model(model, test_loader, criterion)
7. 可视化预测结果
使用以下Python代码来可视化模型的预测结果。
import matplotlib.pyplot as plt
def visualize_predictions(model, test_loader, num_samples=5):
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
model.to(device)
model.eval()
fig, axes = plt.subplots(num_samples, 3, figsize=(15, 5 * num_samples))
with torch.no_grad():
for i, (images, masks) in enumerate(test_loader):
images, masks = images.to(device), masks.to(device)
outputs = model(images)
predictions = torch.sigmoid(outputs).cpu().numpy()
predictions = (predictions > 0.5).astype(np.uint8)
for j in range(min(num_samples, len(images))):
ax = axes[j] if num_samples > 1 else axes
ax[0].imshow(images[j].cpu().permute(1, 2, 0).numpy())
ax[0].set_title("Input Image")
ax[0].axis('off')
ax[1].imshow(masks[j].cpu().numpy(), cmap='gray')
ax[1].set_title("Ground Truth Mask")
ax[1].axis('off')
ax[2].imshow(predictions[j, 0], cmap='gray')
ax[2].set_title("Predicted Mask")
ax[2].axis('off')
if i == 0:
break
plt.tight_layout()
plt.show()
# 可视化预测结果
visualize_predictions(model, test_loader, num_samples=5)
8. 模型优化
为了进一步优化模型,可以尝试以下方法:
- 调整超参数:使用不同的学习率、批量大小、权重衰减等。
- 使用预训练模型:使用预训练的模型作为初始化权重。
- 增加数据量:通过数据增强或收集更多数据来增加训练集的多样性。
- 模型融合:使用多个模型进行集成学习,提高预测的准确性。
9. 总结
通过以上步骤,你可以成功地使用多种深度学习模型对息肉分割数据集进行训练、评估和可视化