数据挖掘-关联分析频繁模式挖掘Apriori、FP-Growth及Eclat算法的JAVA及C++实现
原文转自:http://blog.csdn.net/yangliuy/article/details/7494983
(update 2012.12.28 关于本项目下载及运行的常见问题 FAQ见 newsgroup18828文本分类器、文本聚类器、关联分析频繁模式挖掘算法的Java实现工程下载及运行FAQ )
一、Apriori算法
Apriori是非常经典的关联分析频繁模式挖掘算法,其思想简明,实现方便,只是效率很低,可以作为频繁模式挖掘的入门算法。其主要特点是
1、k-1项集连接规律:若有两个k-1项集,每个项集保证有序,如果两个k-1项集的前k-
2个项相同,而最后一个项不同,则证明它们是可连接的,可连接生成k项集。
2、反单调性。如果一个项集是频繁的,那么它的所有子集都是频繁的。即若一个项集的
子集不是频繁项集,则该项集肯定也不是频繁项集。
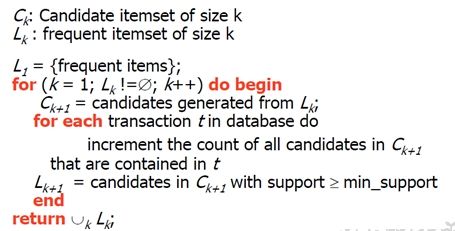
主要算法流程:
1. 扫描数据库,生成候选1项集和频繁1项集。
2. 从2项集开始循环,由频繁k-1项集生成频繁频繁k项集。
2.1 频繁k-1项集两两组合,判定是否可以连接,若能则连接生成k项集。
2.2 对k项集中的每个项集检测其子集是否频繁,舍弃掉子集不是频繁项集即 不在频繁k-1项集中的项集。
2.3 扫描数据库,计算2.3步中过滤后的k项集的支持度,舍弃掉支持度小于阈值的项集,生成频繁k项集。
3. 若当前k项集中只有一个项集时循环结束。
2. 从2项集开始循环,由频繁k-1项集生成频繁频繁k项集。
2.1 频繁k-1项集两两组合,判定是否可以连接,若能则连接生成k项集。
2.2 对k项集中的每个项集检测其子集是否频繁,舍弃掉子集不是频繁项集即 不在频繁k-1项集中的项集。
2.3 扫描数据库,计算2.3步中过滤后的k项集的支持度,舍弃掉支持度小于阈值的项集,生成频繁k项集。
3. 若当前k项集中只有一个项集时循环结束。
伪代码如下:
JAVA实现代码
- package com.pku.yangliu;
- import java.io.BufferedReader;
- import java.io.File;
- import java.io.FileReader;
- import java.io.FileWriter;
- import java.io.IOException;
- import java.util.ArrayList;
- import java.util.HashMap;
- import java.util.HashSet;
- import java.util.List;
- import java.util.Map;
- import java.util.Set;
- import java.util.TreeSet;
- /**频繁模式挖掘算法Apriori实现
- * @author yangliu
- * @qq 772330184
- * @blog http://blog.csdn.net/yangliuy
- * @mail [email protected]
- *
- */
- public class AprioriFPMining {
- private int minSup;//最小支持度
- private static List<Set<String>> dataTrans;//以List<Set<String>>格式保存的事物数据库,利用Set的有序性
- public int getMinSup() {
- return minSup;
- }
- public void setMinSup(int minSup) {
- this.minSup = minSup;
- }
- /**
- * @param args
- */
- public static void main(String[] args) throws IOException {
- AprioriFPMining apriori = new AprioriFPMining();
- double [] threshold = {0.25, 0.20, 0.15, 0.10, 0.05};
- String srcFile = "F:/DataMiningSample/FPmining/Mushroom.dat";
- String shortFileName = srcFile.split("/")[3];
- String targetFile = "F:/DataMiningSample/FPmining/" + shortFileName.substring(0, shortFileName.indexOf("."))+"_fp_threshold";
- dataTrans = apriori.readTrans(srcFile);
- for(int k = 0; k < threshold.length; k++){
- System.out.println(srcFile + " threshold: " + threshold[k]);
- long totalItem = 0;
- long totalTime = 0;
- FileWriter tgFileWriter = new FileWriter(targetFile + (threshold[k]*100));
- apriori.setMinSup((int)(dataTrans.size() * threshold[k]));//原始蘑菇的数据0.25只需要67秒跑出结果
- long startTime = System.currentTimeMillis();
- Map<String, Integer> f1Set = apriori.findFP1Items(dataTrans);
- long endTime = System.currentTimeMillis();
- totalTime += endTime - startTime;
- //频繁1项集信息得加入支持度
- Map<Set<String>, Integer> f1Map = new HashMap<Set<String>, Integer>();
- for(Map.Entry<String, Integer> f1Item : f1Set.entrySet()){
- Set<String> fs = new HashSet<String>();
- fs.add(f1Item.getKey());
- f1Map.put(fs, f1Item.getValue());
- }
- totalItem += apriori.printMap(f1Map, tgFileWriter);
- Map<Set<String>, Integer> result = f1Map;
- do {
- startTime = System.currentTimeMillis();
- result = apriori.genNextKItem(result);
- endTime = System.currentTimeMillis();
- totalTime += endTime - startTime;
- totalItem += apriori.printMap(result, tgFileWriter);
- } while(result.size() != 0);
- tgFileWriter.close();
- System.out.println("共用时:" + totalTime + "ms");
- System.out.println("共有" + totalItem + "项频繁模式");
- }
- }
- /**由频繁K-1项集生成频繁K项集
- * @param preMap 保存频繁K项集的map
- * @param tgFileWriter 输出文件句柄
- * @return int 频繁i项集的数目
- * @throws IOException
- */
- private Map<Set<String>, Integer> genNextKItem(Map<Set<String>, Integer> preMap) {
- // TODO Auto-generated method stub
- Map<Set<String>, Integer> result = new HashMap<Set<String>, Integer>();
- //遍历两个k-1项集生成k项集
- List<Set<String>> preSetArray = new ArrayList<Set<String>>();
- for(Map.Entry<Set<String>, Integer> preMapItem : preMap.entrySet()){
- preSetArray.add(preMapItem.getKey());
- }
- int preSetLength = preSetArray.size();
- for (int i = 0; i < preSetLength - 1; i++) {
- for (int j = i + 1; j < preSetLength; j++) {
- String[] strA1 = preSetArray.get(i).toArray(new String[0]);
- String[] strA2 = preSetArray.get(j).toArray(new String[0]);
- if (isCanLink(strA1, strA2)) { // 判断两个k-1项集是否符合连接成k项集的条件
- Set<String> set = new TreeSet<String>();
- for (String str : strA1) {
- set.add(str);
- }
- set.add((String) strA2[strA2.length - 1]); // 连接成k项集
- // 判断k项集是否需要剪切掉,如果不需要被cut掉,则加入到k项集列表中
- if (!isNeedCut(preMap, set)) {//由于单调性,必须保证k项集的所有k-1项子集都在preMap中出现,否则就该剪切该k项集
- result.put(set, 0);
- }
- }
- }
- }
- return assertFP(result);//遍历事物数据库,求支持度,确保为频繁项集
- }
- /**检测k项集是否该剪切。由于单调性,必须保证k项集的所有k-1项子集都在preMap中出现,否则就该剪切该k项集
- * @param preMap k-1项频繁集map
- * @param set 待检测的k项集
- * @return boolean 是否该剪切
- * @throws IOException
- */
- private boolean isNeedCut(Map<Set<String>, Integer> preMap, Set<String> set) {
- // TODO Auto-generated method stub
- boolean flag = false;
- List<Set<String>> subSets = getSubSets(set);
- for(Set<String> subSet : subSets){
- if(!preMap.containsKey(subSet)){
- flag = true;
- break;
- }
- }
- return flag;
- }
- /**获取k项集set的所有k-1项子集
- * @param set 频繁k项集
- * @return List<Set<String>> 所有k-1项子集容器
- * @throws IOException
- */
- private List<Set<String>> getSubSets(Set<String> set) {
- // TODO Auto-generated method stub
- String[] setArray = set.toArray(new String[0]);
- List<Set<String>> result = new ArrayList<Set<String>>();
- for(int i = 0; i < setArray.length; i++){
- Set<String> subSet = new HashSet<String>();
- for(int j = 0; j < setArray.length; j++){
- if(j != i) subSet.add(setArray[j]);
- }
- result.add(subSet);
- }
- return result;
- }
- /**遍历事物数据库,求支持度,确保为频繁项集
- * @param allKItem 候选频繁k项集
- * @return Map<Set<String>, Integer> 支持度大于阈值的频繁项集和支持度map
- * @throws IOException
- */
- private Map<Set<String>, Integer> assertFP(
- Map<Set<String>, Integer> allKItem) {
- // TODO Auto-generated method stub
- Map<Set<String>, Integer> result = new HashMap<Set<String>, Integer>();
- for(Set<String> kItem : allKItem.keySet()){
- for(Set<String> data : dataTrans){
- boolean flag = true;
- for(String str : kItem){
- if(!data.contains(str)){
- flag = false;
- break;
- }
- }
- if(flag) allKItem.put(kItem, allKItem.get(kItem) + 1);
- }
- if(allKItem.get(kItem) >= minSup) {
- result.put(kItem, allKItem.get(kItem));
- }
- }
- return result;
- }
- /**检测两个频繁K项集是否可以连接,连接条件是只有最后一个项不同
- * @param strA1 k项集1
- * @param strA1 k项集2
- * @return boolean 是否可以连接
- * @throws IOException
- */
- private boolean isCanLink(String[] strA1, String[] strA2) {
- // TODO Auto-generated method stub
- boolean flag = true;
- if(strA1.length != strA2.length){
- return false;
- }else {
- for(int i = 0; i < strA1.length - 1; i++){
- if(!strA1[i].equals(strA2[i])){
- flag = false;
- break;
- }
- }
- if(strA1[strA1.length -1].equals(strA2[strA1.length -1])){
- flag = false;
- }
- }
- return flag;
- }
- /**将频繁i项集的内容及支持度输出到文件 格式为 模式:支持度
- * @param f1Map 保存频繁i项集的容器<i项集 , 支持度>
- * @param tgFileWriter 输出文件句柄
- * @return int 频繁i项集的数目
- * @throws IOException
- */
- private int printMap(Map<Set<String>, Integer> f1Map, FileWriter tgFileWriter) throws IOException {
- // TODO Auto-generated method stub
- for(Map.Entry<Set<String>, Integer> f1MapItem : f1Map.entrySet()){
- for(String p : f1MapItem.getKey()){
- tgFileWriter.append(p + " ");
- }
- tgFileWriter.append(": " + f1MapItem.getValue() + "\n");
- }
- tgFileWriter.flush();
- return f1Map.size();
- }
- /**生成频繁1项集
- * @param fileDir 事务文件目录
- * @return Map<String, Integer> 保存频繁1项集的容器<1项集 , 支持度>
- * @throws IOException
- */
- private Map<String, Integer> findFP1Items(List<Set<String>> dataTrans) {
- // TODO Auto-generated method stub
- Map<String, Integer> result = new HashMap<String, Integer>();
- Map<String, Integer> itemCount = new HashMap<String, Integer>();
- for(Set<String> ds : dataTrans){
- for(String d : ds){
- if(itemCount.containsKey(d)){
- itemCount.put(d, itemCount.get(d) + 1);
- } else {
- itemCount.put(d, 1);
- }
- }
- }
- for(Map.Entry<String, Integer> ic : itemCount.entrySet()){
- if(ic.getValue() >= minSup){
- result.put(ic.getKey(), ic.getValue());
- }
- }
- return result;
- }
- /**读取事务数据库
- * @param fileDir 事务文件目录
- * @return List<String> 保存事务的容器
- * @throws IOException
- */
- private List<Set<String>> readTrans(String fileDir) {
- // TODO Auto-generated method stub
- List<Set<String>> records = new ArrayList<Set<String>>();
- try {
- FileReader fr = new FileReader(new File(fileDir));
- BufferedReader br = new BufferedReader(fr);
- String line = null;
- while ((line = br.readLine()) != null) {
- if (line.trim() != "") {
- Set<String> record = new HashSet<String>();
- String[] items = line.split(" ");
- for (String item : items) {
- record.add(item);
- }
- records.add(record);
- }
- }
- } catch (IOException e) {
- System.out.println("读取事务文件失败。");
- System.exit(-2);
- }
- return records;
- }
- }
实验结果
F:/DataMiningSample/FPmining/Mushroom.dat threshold: 0.25
共用时:54015ms
共有5545项频繁模式
F:/DataMiningSample/FPmining/Mushroom.dat threshold: 0.2
共用时:991610ms
共有53663项频繁模式
F:/DataMiningSample/FPmining/Mushroom.dat threshold: 0.15
结论:对Mushroom.dat挖掘出来的频繁模式及支持度、频繁模式总数正确,但是算法速度很慢,对大数据量如T10I4D100K低阈值挖掘时间太长
解决办法:改用C++写FP-Growth算法做频繁模式挖掘!
解决办法:改用C++写FP-Growth算法做频繁模式挖掘!
二、FP-Growth算法
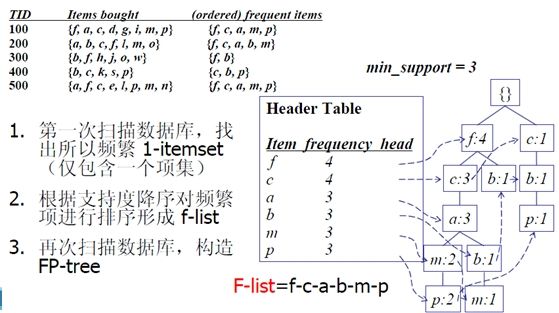
FP-Growth算法由数据挖掘界大牛Han Jiawei教授于SIGMOD 00‘大会提出,提出根据事物数据库构建FP-Tree,然后基于FP-Tree生成频繁模式集。主要算法流程如下
Step1 读取数据库,构造频繁1项集及FP-tree
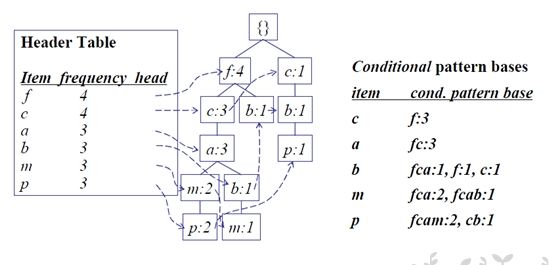
Step3 对CPB上每一条路径的节点更新计数为x的计数,根据CPB构造条件FP-tree
Step4 从条件FP-tree中找到所有长路径,对该路径上的节点找出所有组合方式,然后合并计数
Step5 将Step4中的频繁项集与x合并,得到包含x的频繁项集
Step2-5 循环,直到遍历头表中的所有项
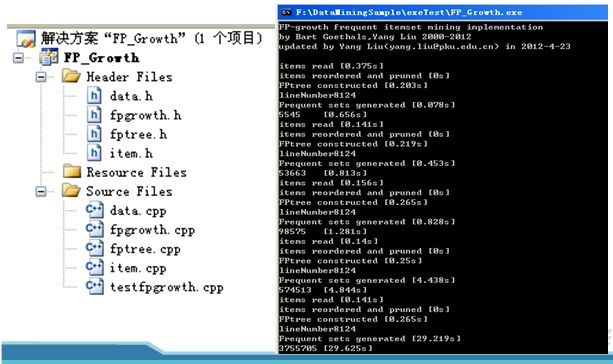
由于时间关系,主要基于芬兰教授Bart Goethals的开源代码实现,源码下载见点击打开链接 ,文件结构及运行结果如下
对Mushroom.dat,accidents.dat和T10I4D100K.dat三个数据集做频繁模式挖掘的结果如下
三、Eclat算法
Eclat算法加入了倒排的思想,加快频繁集生成速度,其算法思想是 由频繁k项集求交集,生成候选k+1项集 。对候选k+1项集做裁剪,生成频繁k+1项集,再求交集生成候选k+2项集。如此迭代,直到项集归一。
算法过程:
1.一次扫描数据库,获得初始数据。包括频繁1项集,数据库包含的所有items,事务总数(行)transNum,最小支持度minsup=limitValue*trans。
2.二次扫描数据库,获得频繁2项集。
3.按照Eclat算法,对频繁2项集迭代求交集,做裁剪,直到项集归一。
Eclat算法加入了倒排的思想,加快频繁集生成速度,其算法思想是 由频繁k项集求交集,生成候选k+1项集 。对候选k+1项集做裁剪,生成频繁k+1项集,再求交集生成候选k+2项集。如此迭代,直到项集归一。
算法过程:
1.一次扫描数据库,获得初始数据。包括频繁1项集,数据库包含的所有items,事务总数(行)transNum,最小支持度minsup=limitValue*trans。
2.二次扫描数据库,获得频繁2项集。
3.按照Eclat算法,对频繁2项集迭代求交集,做裁剪,直到项集归一。
JAVA实现如下
- package com.pku.yhf;
- import java.io.BufferedReader;
- import java.io.BufferedWriter;
- import java.io.File;
- import java.io.FileInputStream;
- import java.io.FileNotFoundException;
- import java.io.FileReader;
- import java.io.FileWriter;
- import java.io.IOException;
- import java.io.InputStreamReader;
- import java.util.ArrayList;
- import java.util.BitSet;
- import java.util.Iterator;
- import java.util.Set;
- import java.util.TreeMap;
- import java.util.TreeSet;
- public class EclatRelease {
- private File file=new File("D:/mushroom.dat.txt");
- private float limitValue=0.25f;
- private int transNum=0;
- private ArrayList<HeadNode> array=new ArrayList<HeadNode>();
- private HashHeadNode[] hashTable;//存放临时生成的频繁项集,作为重复查询的备选集合
- public long newItemNum=0;
- private File tempFile=null;
- private BufferedWriter bw=null;
- public static long modSum=0;
- /**
- * 第一遍扫描数据库,确定Itemset,根据阈值计算出支持度数
- */
- public void init()
- {
- Set itemSet=new TreeSet();
- MyMap<Integer,Integer> itemMap=new MyMap<Integer,Integer>();
- int itemNum=0;
- Set[][] a;
- try {
- FileInputStream fis=new FileInputStream(file);
- BufferedReader br=new BufferedReader(new InputStreamReader(fis));
- String str=null;
- //第一次扫描数据集合
- while((str=br.readLine()) != null)
- {
- transNum++;
- String[] line=str.split(" ");
- for(String item:line)
- {
- itemSet.add(Integer.parseInt(item));
- itemMap.add(Integer.parseInt((item)));
- }
- }
- br.close();
- // System.out.println("itemMap lastKey:"+itemMap.lastKey());
- // System.out.println("itemsize:"+itemSet.size());
- // System.out.println("trans: "+transNum);
- //ItemSet.limitSupport=(int)Math.ceil(transNum*limitValue);//上取整
- ItemSet.limitSupport=(int)Math.floor(transNum*limitValue);//下取整
- ItemSet.ItemSize=(Integer)itemMap.lastKey();
- ItemSet.TransSize=transNum;
- hashTable=new HashHeadNode[ItemSet.ItemSize*3];//生成项集hash表
- for(int i=0;i<hashTable.length;i++)
- {
- hashTable[i]=new HashHeadNode();
- }
- // System.out.println("limitSupport:"+ItemSet.limitSupport);
- tempFile=new File(file.getParent()+"/"+file.getName()+".dat");
- if(tempFile.exists())
- {
- tempFile.delete();
- }
- tempFile.createNewFile();
- bw=new BufferedWriter(new FileWriter(tempFile));
- Set oneItem=itemMap.keySet();
- int countOneItem=0;
- for(Iterator it=oneItem.iterator();it.hasNext();)
- {
- int key=(Integer)it.next();
- int value=(Integer)itemMap.get(key);
- if(value >= ItemSet.limitSupport)
- {
- bw.write(key+" "+":"+" "+value);
- bw.write("\n");
- countOneItem++;
- }
- }
- bw.flush();
- modSum+=countOneItem;
- itemNum=(Integer)itemMap.lastKey();
- a=new TreeSet[itemNum+1][itemNum+1];
- array.add(new HeadNode());//空项
- for(short i=1;i<=itemNum;i++)
- {
- HeadNode hn=new HeadNode();
- // hn.item=i;
- array.add(hn);
- }
- BufferedReader br2=new BufferedReader(new FileReader(file));
- //第二次扫描数据集合,形成2-项候选集
- int counter=0;//事务
- int max=0;
- while((str=br2.readLine()) != null)
- {max++;
- String[] line=str.split(" ");
- counter++;
- for(int i=0;i<line.length;i++)
- {
- int sOne=Integer.parseInt(line[i]);
- for(int j=i+1;j<line.length;j++)
- {
- int sTwo=Integer.parseInt(line[j]);
- if(a[sOne][sTwo] == null)
- {
- Set set=new TreeSet();
- set.add(counter);
- a[sOne][sTwo]=set;
- }
- else{
- a[sOne][sTwo].add(counter);
- }
- }
- }
- }
- //将数组集合转换为链表集合
- for(int i=1;i<=itemNum;i++)
- {
- HeadNode hn=array.get(i);
- for(int j=i+1;j<=itemNum;j++)
- {
- if(a[i][j] != null && a[i][j].size() >= ItemSet.limitSupport)
- {
- hn.items++;
- ItemSet is=new ItemSet(true);
- is.item=2;
- is.items.set(i);
- is.items.set(j);
- is.supports=a[i][j].size();
- bw.write(i+" "+j+" "+": "+is.supports);
- bw.write("\n");
- //统计频繁2-项集的个数
- modSum++;
- for(Iterator it=a[i][j].iterator();it.hasNext();)
- {
- int value=(Integer)it.next();
- is.trans.set(value);
- }
- if( hn.first== null)
- {
- hn.first=is;
- hn.last=is;
- }
- else{
- hn.last.next=is;
- hn.last=is;
- }
- }
- }
- }
- bw.flush();
- } catch (FileNotFoundException e) {
- e.printStackTrace();
- } catch (IOException e) {
- e.printStackTrace();
- }
- }
- public void start()
- {
- boolean flag=true;
- //TreeSet ts=new TreeSet();//临时存储项目集合,防止重复项集出现,节省空间
- int count=0;
- ItemSet shareFirst=new ItemSet(false);
- while(flag)
- {
- flag=false;
- //System.out.println(++count);
- for(int i=1;i<array.size();i++)
- {
- HeadNode hn=array.get(i);
- if(hn.items > 1 )//项集个数大于1
- {
- generateLargeItemSet(hn,shareFirst);
- flag=true;
- }
- clear(hashTable);
- }
- }try {
- bw.close();
- } catch (IOException e) {
- e.printStackTrace();
- }
- }
- public void generateLargeItemSet(HeadNode hn,ItemSet shareFirst){
- BitSet bsItems=new BitSet(ItemSet.ItemSize);//存放链两个k-1频繁项集的ItemSet交
- BitSet bsTrans=new BitSet(ItemSet.TransSize);//存放两个k-1频繁项集的Trans交
- BitSet containItems=new BitSet(ItemSet.ItemSize);//存放两个k-1频繁项集的ItemSet的并
- BitSet bsItems2=new BitSet(ItemSet.ItemSize);//临时存放容器BitSet
- ItemSet oldCurrent=null,oldNext=null;
- oldCurrent=hn.first;
- long countItems=0;
- ItemSet newFirst=new ItemSet(false),newLast=newFirst;
- while(oldCurrent != null)
- {
- oldNext=oldCurrent.next;
- while(oldNext != null)
- {
- //生成k—项候选集,由两个k-1项频繁集生成
- bsItems.clear();
- bsItems.or(oldCurrent.items);
- bsItems.and(oldNext.items);
- if(bsItems.cardinality() < oldCurrent.item-1)
- {
- break;
- }
- //新合并的项集是否已经存在
- containItems.clear();
- containItems.or(oldCurrent.items);//将k-1项集合并
- containItems.or(oldNext.items);
- if(!containItems(containItems,bsItems2,newFirst)){
- bsTrans.clear();
- bsTrans.or(oldCurrent.trans);
- bsTrans.and(oldNext.trans);
- if(bsTrans.cardinality() >= ItemSet.limitSupport)
- {
- ItemSet is=null;
- if(shareFirst.next == null)//没有共享ItemSet链表
- {
- is=new ItemSet(true);
- newItemNum++;
- }
- else
- {
- is=shareFirst.next;
- shareFirst.next=shareFirst.next.next;
- is.items.clear();
- is.trans.clear();
- is.next=null;
- }
- is.item=(oldCurrent.item+1);//生成k—项候选集,由两个k-1项频繁集生成
- is.items.or(oldCurrent.items);//将k-1项集合并
- is.items.or(oldNext.items);//将k-1项集合并
- is.trans.or(oldCurrent.trans);//将bs1的值复制到bs中
- is.trans.and(oldNext.trans);
- is.supports=is.trans.cardinality();
- writeToFile(is.items,is.supports);//将频繁项集及其支持度写入文件
- countItems++;
- modSum++;
- newLast.next=is;
- newLast=is;
- }
- }
- oldNext=oldNext.next;
- }
- oldCurrent=oldCurrent.next;
- }
- ItemSet temp1=hn.first;
- ItemSet temp2=hn.last;
- temp2.next=shareFirst.next;
- shareFirst.next=temp1;
- hn.first=newFirst.next;
- hn.last=newLast;
- hn.items=countItems;
- }
- public boolean containItems(BitSet containItems,BitSet bsItems2,ItemSet first)
- {
- long size=containItems.cardinality();//项集数目
- int itemSum=0;
- int temp=containItems.nextSetBit(0);
- while(true)
- {
- itemSum+=temp;
- temp=containItems.nextSetBit(temp+1);
- if(temp == -1)
- {
- break;
- }
- }
- int hash=itemSum%(ItemSet.ItemSize*3);
- HashNode hn=hashTable[hash].next;
- Node pre=hashTable[hash];
- while(true)
- {
- if(hn == null)//不包含containItems
- {
- HashNode node=new HashNode();
- node.bs.or(containItems);
- pre.next=node;
- return false;
- }
- if(hn.bs.isEmpty())
- {
- hn.bs.or(containItems);
- return false;
- }
- bsItems2.clear();
- bsItems2.or(containItems);
- bsItems2.and(hn.bs);
- if(bsItems2.cardinality() == size)
- {
- return true;
- }
- pre=hn;
- hn=hn.next;
- }
- }
- public void clear(HashHeadNode[] hashTable)
- {
- for(int i=0;i<hashTable.length;i++)
- {
- HashNode node=hashTable[i].next;
- while(node != null)
- {
- node.bs.clear();
- node=node.next;
- }
- }
- }
- public void writeToFile(BitSet items,int supports)
- {
- StringBuilder sb=new StringBuilder();
- //sb.append("<");
- int temp=items.nextSetBit(0);
- sb.append(temp);
- while(true)
- {
- temp=items.nextSetBit(temp+1);
- if(temp == -1)
- {
- break;
- }
- //sb.append(",");
- sb.append(" ");
- sb.append(temp);
- }
- sb.append(" :"+" "+supports);
- try {
- bw.write(sb.toString());
- bw.write("\n");
- } catch (IOException e) {
- e.printStackTrace();
- }
- }
- public static void main(String[] args) {
- EclatRelease e=new EclatRelease();
- long begin=System.currentTimeMillis();
- e.init();
- e.start();
- long end=System.currentTimeMillis();
- double time=(double)(end-begin)/1000;
- System.out.println("共耗时"+time+"秒");
- System.out.println("频繁模式数目:"+EclatRelease.modSum);
- }
- }
- class MyMap<T,E> extends TreeMap
- {
- public void add(T obj)
- {
- if(this.containsKey(obj))
- {
- int value=(Integer)this.get(obj);
- this.put(obj, value+1);
- }
- else
- this.put(obj, 1);
- }
- }
- package com.pku.yhf;
- import java.util.BitSet;
- public class ItemSet {
- public static int limitSupport;//根据阈值计算出的最小支持度数
- public static int ItemSize;//Items数目
- public static int TransSize; //事务数目
- public boolean flag=true; //true,表示作为真正的ItemSet,false只作为标记节点,只在HashTabel中使用
- public int item=0;// 某项集
- public int supports=0;//项集的支持度
- public BitSet items=null;
- public BitSet trans=null;
- //public TreeSet items=new TreeSet();//项集
- //public TreeSet trans=new TreeSet();//事务集合
- public ItemSet next=null;//下一个项集
- public ItemSet(boolean flag)
- {
- this.flag=flag;
- if(flag)
- {
- item=0;// 某项集
- supports=0;//项集的支持度
- items=new BitSet(ItemSize+1);
- trans=new BitSet(TransSize+1);
- }
- }
- }
对mushroom.dat的频繁模式挖掘结果如下