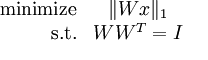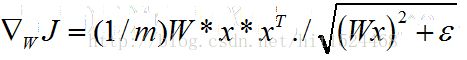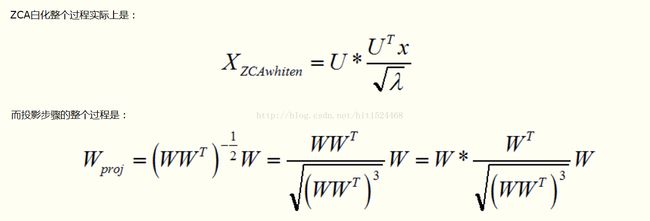独立成分分析及Demo
1. 独立成分分析建模
独立成分分析目标是实现在海量数据中学习完备的单位正交基,所以,所以,可以建立如下的最优化问题:
其中,第一项为稀疏约束,第二项为完备单位正交基约束,熟悉稀疏表示的可能会注意到,为什么上述最优化问题中,没有显式加入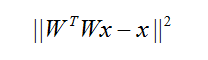 约束,因为W为正交矩阵,所以上述误差为0.
约束,因为W为正交矩阵,所以上述误差为0.
2. 独立成分分析最优化问题求解
(1)稀疏约束优化:转化为如下能量泛函的最优化问题
其导数如下:
(2)完备单位正交基约束优化:通过投影实现,投影如下:
注意,由于我们的完备单位正交基来源于数据,所以,在进行迭代训练前,需要对数据样本进行ZCA White和投影的对比如下:
(3) 优化步骤:
step1: 对训练数据进行中心化、ZCA White处理
step2:根据梯度下降法更新W
step3:对W进行投影
step4:是否满足收敛条件,满足则结束,否则重复step2-3;
(4) 示例代码:
%% CS294A/CS294W Independent Component Analysis (ICA) Exercise
% Instructions
% ------------
%
% This file contains code that helps you get started on the
% ICA exercise. In this exercise, you will need to modify
% orthonormalICACost.m and a small part of this file, ICAExercise.m.
%%======================================================================
%% STEP 0: Initialization
% Here we initialize some parameters used for the exercise.
numPatches = 20000;
numFeatures = 121;
imageChannels = 3;
patchDim = 8;
visibleSize = patchDim * patchDim * imageChannels;
outputDir = '.';
epsilon = 1e-6; % L1-regularisation epsilon |Wx| ~ sqrt((Wx).^2 + epsilon)
%%======================================================================
%% STEP 1: Sample patches
patches = load('stlSampledPatches.mat');
patches = patches.patches(:, 1:numPatches);
displayColorNetwork(patches(:, 1:100));
%%======================================================================
%% STEP 2: ZCA whiten patches
% In this step, we ZCA whiten the sampled patches. This is necessary for
% orthonormal ICA to work.
patches = patches / 255;
meanPatch = mean(patches, 2);
patches = bsxfun(@minus, patches, meanPatch);
% 数据进行ZCA White处理
sigma = patches * patches';
[u, s, v] = svd(sigma);
ZCAWhite = u * diag(1 ./ sqrt(diag(s))) * u';
patches = ZCAWhite * patches;
%%======================================================================
%% STEP 3: ICA cost functions
% Implement the cost function for orthornomal ICA (you don't have to
% enforce the orthonormality constraint in the cost function)
% in the function orthonormalICACost in orthonormalICACost.m.
% Once you have implemented the function, check the gradient.
% Use less features and smaller patches for speed
debug = false; % 置为true时,进行梯度计算验证
if debug
numFeatures = 5;
patches = patches(1:3, 1:5);
visibleSize = 3;
numPatches = 5;
weightMatrix = rand(numFeatures, visibleSize);
[cost, grad] = orthonormalICACost(weightMatrix, visibleSize, numFeatures, patches, epsilon);
numGrad = computeNumericalGradient( @(x) orthonormalICACost(x, visibleSize, numFeatures, patches, epsilon), weightMatrix(:) );
% Uncomment to display the numeric and analytic gradients side-by-side
% disp([numGrad grad]);
diff = norm(numGrad-grad)/norm(numGrad+grad);
fprintf('Orthonormal ICA difference: %g\n', diff);
assert(diff < 1e-7, 'Difference too large. Check your analytic gradients.');
fprintf('Congratulations! Your gradients seem okay.\n');
end
%%======================================================================
%% STEP 4: Optimization for orthonormal ICA
% Optimize for the orthonormal ICA objective, enforcing the orthonormality
% constraint. Code has been provided to do the gradient descent with a
% backtracking line search using the orthonormalICACost function
% (for more information about backtracking line search, you can read the
% appendix of the exercise).
%
% However, you will need to write code to enforce the orthonormality
% constraint by projecting weightMatrix back into the space of matrices
% satisfying WW^T = I.
%
% Once you are done, you can run the code. 10000 iterations of gradient
% descent will take around 2 hours, and only a few bases will be
% completely learned within 10000 iterations. This highlights one of the
% weaknesses of orthonormal ICA - it is difficult to optimize for the
% objective function while enforcing the orthonormality constraint -
% convergence using gradient descent and projection is very slow.
weightMatrix = rand(numFeatures, visibleSize);
[cost, grad] = orthonormalICACost(weightMatrix(:), visibleSize, numFeatures, patches, epsilon);% 计算梯度
fprintf('%11s%16s%10s\n','Iteration','Cost','t');
startTime = tic();
% Initialize some parameters for the backtracking line search
alpha = 0.5;
t = 0.02;
lastCost = 1e40;
% Do 10000 iterations of gradient descent
for iteration = 1:50000
grad = reshape(grad, size(weightMatrix));
newCost = Inf;
linearDelta = sum(sum(grad .* grad));
% Perform the backtracking line search
while 1
considerWeightMatrix = weightMatrix - alpha * grad; %梯度下降法更新W
% -------------------- YOUR CODE HERE --------------------
% Instructions:
% Write code to project considerWeightMatrix back into the space
% of matrices satisfying WW^T = I.
%
% Once that is done, verify that your projection is correct by
% using the checking code below. After you have verified your
% code, comment out the checking code before running the
% optimization.
% % Project considerWeightMatrix such that it satisfies WW^T = I
% error('Fill in the code for the projection here');
considerWeightMatrix = (considerWeightMatrix*considerWeightMatrix')^(-0.5)*considerWeightMatrix; % W进行投影
% Verify that the projection is correct
temp = considerWeightMatrix * considerWeightMatrix';
temp = temp - eye(numFeatures);
assert(sum(temp(:).^2) < 1e-23, 'considerWeightMatrix does not satisfy WW^T = I. Check your projection again');
% error('Projection seems okay. Comment out verification code before running optimization.');
% -------------------- YOUR CODE HERE --------------------
[newCost, newGrad] = orthonormalICACost(considerWeightMatrix(:), visibleSize, numFeatures, patches, epsilon);
if newCost > lastCost - alpha * t * linearDelta
% fprintf(' %14.6f %14.6f\n', newCost, lastCost - alpha * t * linearDelta);
t = 0.9 * t;
else
break;
end
end
lastCost = newCost;
weightMatrix = considerWeightMatrix;
fprintf(' %9d %14.6f %8.7g\n', iteration, newCost, t);
t = 1.1 * t;
cost = newCost;
grad = newGrad;
% Visualize the learned bases as we go along
if mod(iteration, 1000) == 0
duration = toc(startTime);
% Visualize the learned bases over time in different figures so
% we can get a feel for the slow rate of convergence
figure(floor(iteration / 1000));
displayColorNetwork(weightMatrix');
end
end
% Visualize the learned bases
displayColorNetwork(weightMatrix');% function [cost, grad] = orthonormalICACost(theta, visibleSize, numFeatures, patches, epsilon)
% %orthonormalICACost - compute the cost and gradients for orthonormal ICA
% % (i.e. compute the cost ||Wx||_1 and its gradient)
%
% weightMatrix = reshape(theta, numFeatures, visibleSize);
%
% cost = 0;
% grad = zeros(numFeatures, visibleSize);
%
% % -------------------- YOUR CODE HERE --------------------
% % Instructions:
% % Write code to compute the cost and gradient with respect to the
% % weights given in weightMatrix.
% % -------------------- YOUR CODE HERE --------------------
% num_samples = size(patches,2); %样本个数
%
% aux1 = sqrt(((weightMatrix*patches).^2) + epsilon);
% cost = sum(aux1(:))/num_samples;
% grad = ((weightMatrix*patches)./aux1)*patches'./num_samples;
% grad = grad(:);
%
% end
%
function [cost, grad] = orthonormalICACost(theta, visibleSize, numFeatures, patches, epsilon)
%orthonormalICACost - compute the cost and gradients for orthonormal ICA
% (i.e. compute the cost ||Wx||_1 and its gradient)
weightMatrix = reshape(theta, numFeatures, visibleSize);
cost = 0;
grad = zeros(numFeatures, visibleSize);
% -------------------- YOUR CODE HERE --------------------
% Instructions:
% Write code to compute the cost and gradient with respect to the
% weights given in weightMatrix.
% -------------------- YOUR CODE HERE --------------------
%% 方法
lambda = 8e-6;% 0.5e-4
num_samples = size(patches,2);
cost_part1 = sum(sum((weightMatrix'*weightMatrix*patches-patches).^2))./num_samples;
cost_part2 = sum(sum(sqrt((weightMatrix*patches).^2+epsilon)))*lambda;
cost = cost_part1 + cost_part2;
grad = (2*weightMatrix*(weightMatrix'*weightMatrix*patches-patches)*patches'+...
2*weightMatrix*patches*(weightMatrix'*weightMatrix*patches-patches)')./num_samples+...
(weightMatrix*patches./sqrt((weightMatrix*patches).^2+epsilon))*patches'*lambda;
grad = grad(:);
fprintf('%11s%16s\n','cost_part1','cost_part2');
fprintf(' %14.6f %14.6f\n', cost_part1, cost_part2);
end注意:计算梯度时,将||w'wx-x||^2的带入求导了!
function numgrad = computeNumericalGradient(J, theta)
% numgrad = computeNumericalGradient(J, theta)
% theta: a vector of parameters
% J: a function that outputs a real-number. Calling y = J(theta) will return the
% function value at theta.
% Initialize numgrad with zeros
numgrad = zeros(size(theta));
%% ---------- YOUR CODE HERE --------------------------------------
% Instructions:
% Implement numerical gradient checking, and return the result in numgrad.
% (See Section 2.3 of the lecture notes.)
% You should write code so that numgrad(i) is (the numerical approximation to) the
% partial derivative of J with respect to the i-th input argument, evaluated at theta.
% I.e., numgrad(i) should be the (approximately) the partial derivative of J with
% respect to theta(i).
%
% Hint: You will probably want to compute the elements of numgrad one at a time.
epsilon = 1e-8;
n = size(theta,1);
E = eye(n);
for i = 1:n
delta = E(:,i)*epsilon;
numgrad(i) = (J(theta+delta)-J(theta-delta))/(epsilon*2.0);
end
%% ---------------------------------------------------------------
end
function displayColorNetwork(A)
% display receptive field(s) or basis vector(s) for image patches
%
% A the basis, with patches as column vectors
% In case the midpoint is not set at 0, we shift it dynamically
if min(A(:)) >= 0
A = A - mean(A(:));
end
cols = round(sqrt(size(A, 2)));
channel_size = size(A,1) / 3;
dim = sqrt(channel_size);
dimp = dim+1;
rows = ceil(size(A,2)/cols);
B = A(1:channel_size,:);
C = A(channel_size+1:channel_size*2,:);
D = A(2*channel_size+1:channel_size*3,:);
B=B./(ones(size(B,1),1)*max(abs(B)));
C=C./(ones(size(C,1),1)*max(abs(C)));
D=D./(ones(size(D,1),1)*max(abs(D)));
% Initialization of the image
I = ones(dim*rows+rows-1,dim*cols+cols-1,3);
%Transfer features to this image matrix
for i=0:rows-1
for j=0:cols-1
if i*cols+j+1 > size(B, 2)
break
end
% This sets the patch
I(i*dimp+1:i*dimp+dim,j*dimp+1:j*dimp+dim,1) = ...
reshape(B(:,i*cols+j+1),[dim dim]);
I(i*dimp+1:i*dimp+dim,j*dimp+1:j*dimp+dim,2) = ...
reshape(C(:,i*cols+j+1),[dim dim]);
I(i*dimp+1:i*dimp+dim,j*dimp+1:j*dimp+dim,3) = ...
reshape(D(:,i*cols+j+1),[dim dim]);
end
end
I = I + 1;
I = I / 2;
imagesc(I);
axis equal
axis off
end
显示前100个样本如下:
迭代10000次时,训练获得的完备单位正交基
参考:
http://www.cnblogs.com/dmzhuo/p/5009274.html
http://deeplearning.stanford.edu/wiki/index.php/Exercise:Independent_Component_Analysis