from keras import layers
from keras.models import Model
import keras.backend as K

K.clear_session()
densenet = DenseNet121(weights=None,include_top=False,input_shape=(None,None,3))
GAP_layer = layers.GlobalAveragePooling2D()
drop_layer = layers.Dropout(0.5)
dense_layer = layers.Dense(5, activation='sigmoid', name='final_output')
def build_model_sequential():
model = Sequential()
model.add(densenet)
model.add(GAP_layer)
model.add(drop_layer)
model.add(dense_layer)
return model
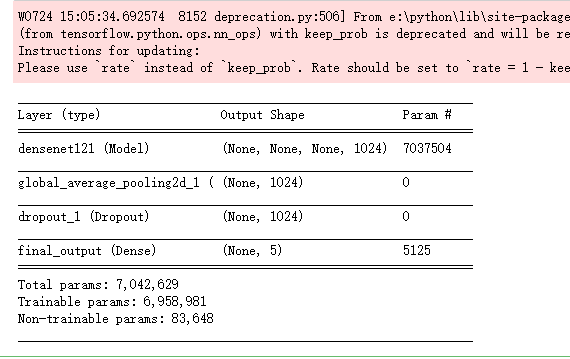
modelA = build_model_sequential()
modelA.load_weights('F:\\kaggleDataSet\\diabeticRetinopathy\\dense_xhlulu_731.h5')
modelA.summary()
def build_model_functional():
base_model = densenet
x = GAP_layer(base_model.layers[-1].output)
x = drop_layer(x)
final_output = dense_layer(x)
model = Model(base_model.layers[0].input, final_output)
return model
model = build_model_functional() # with pretrained weights, and layers we want
model.summary()

y_test = model.predict(x_test) > 0.5
y_test = y_test.astype(int).sum(axis=1) - 1
import seaborn as sns
import cv2
SIZE=224
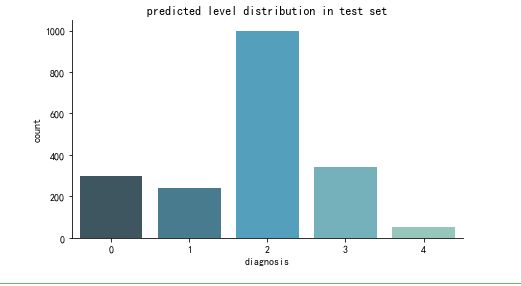
def create_pred_hist(pred_level_y,title='NoTitle'):
results = pd.DataFrame({'diagnosis':pred_level_y})
f, ax = plt.subplots(figsize=(7, 4))
ax = sns.countplot(x="diagnosis", data=results, palette="GnBu_d")
sns.despine()
plt.title(title)
plt.show()
create_pred_hist(y_test,title='predicted level distribution in test set')
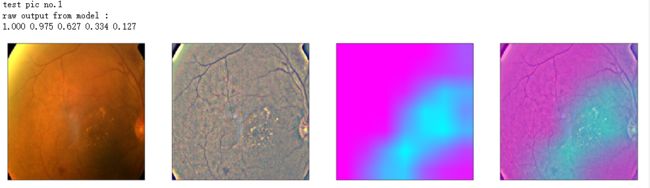
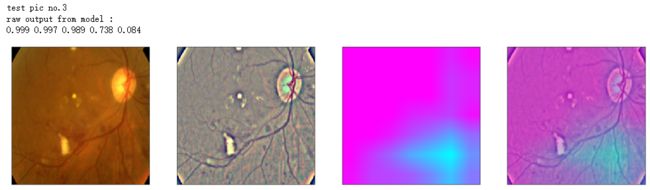
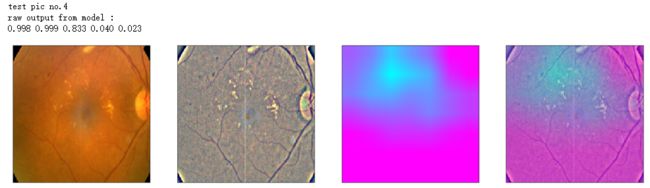
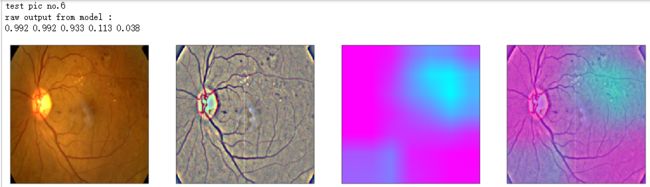
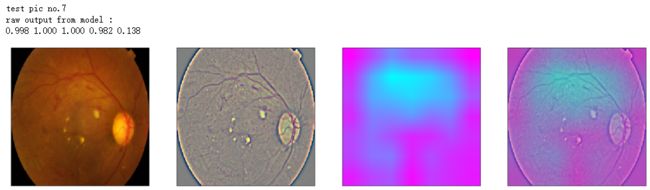
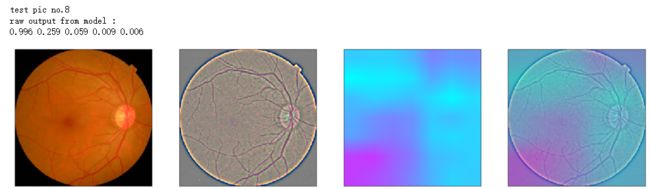
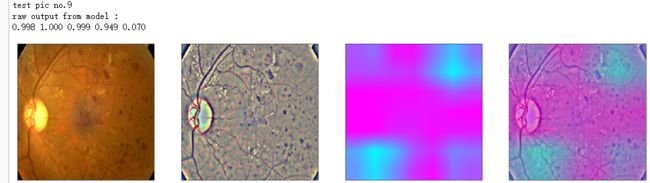
def gen_heatmap_img(img, model0, layer_name='last_conv_layer',viz_img=None,orig_img=None):
preds_raw = model0.predict(img[np.newaxis])
preds = preds_raw > 0.5 # use the same threshold as @xhlulu original kernel
class_idx = (preds.astype(int).sum(axis=1) - 1)[0]
class_output_tensor = model0.output[:, class_idx]
viz_layer = model0.get_layer(layer_name)
grads = K.gradients(class_output_tensor ,viz_layer.output)[0] # gradients of viz_layer wrt output_tensor of predicted class
pooled_grads=K.mean(grads,axis=(0,1,2))
iterate=K.function([model0.input],[pooled_grads, viz_layer.output[0]])
pooled_grad_value, viz_layer_out_value = iterate([img[np.newaxis]])
for i in range(pooled_grad_value.shape[0]):
viz_layer_out_value[:,:,i] *= pooled_grad_value[i]
heatmap = np.mean(viz_layer_out_value, axis=-1)
heatmap = np.maximum(heatmap,0)
heatmap /= np.max(heatmap)
viz_img=cv2.resize(viz_img,(img.shape[1],img.shape[0]))
heatmap=cv2.resize(heatmap,(viz_img.shape[1],viz_img.shape[0]))
heatmap_color = cv2.applyColorMap(np.uint8(heatmap*255), cv2.COLORMAP_SPRING)/255
heated_img = heatmap_color*0.5 + viz_img*0.5
print('raw output from model : ')
print_pred(preds_raw)
if orig_img is None:
show_Nimages([img,viz_img,heatmap_color,heated_img])
else:
show_Nimages([orig_img,img,viz_img,heatmap_color,heated_img])
plt.show()
return heated_img
def show_image(image,figsize=None,title=None):
if figsize is not None:
fig = plt.figure(figsize=figsize)
if image.ndim == 2:
plt.imshow(image,cmap='gray')
else:
plt.imshow(image)
if title is not None:
plt.title(title)
def show_Nimages(imgs,scale=1):
N=len(imgs)
fig = plt.figure(figsize=(25/scale, 16/scale))
for i, img in enumerate(imgs):
ax = fig.add_subplot(1, N, i + 1, xticks=[], yticks=[])
show_image(img)
def print_pred(array_of_classes):
xx = array_of_classes
s1,s2 = xx.shape
for i in range(s1):
for j in range(s2):
print('%.3f ' % xx[i,j],end='')
print('')
NUM_SAMP=10
SEED=77
layer_name = 'relu' #'conv5_block16_concat'
for i, (idx, row) in enumerate(test_df[:NUM_SAMP].iterrows()):
path=f"F:\\kaggleDataSet\\diabeticRetinopathy\\resized test 19\\"+str(image_id)+".jpg"
ben_img = load_image_ben_orig(path)
input_img = np.empty((1,224, 224, 3), dtype=np.uint8)
input_img[0,:,:,:] = preprocess_image(path)
print('test pic no.%d' % (i+1))
_ = gen_heatmap_img(input_img[0],model, layer_name=layer_name,viz_img=ben_img)