Course 2 改善深层神经网络 Week 2 mini-batch梯度下降法、momentum梯度下降和Adam优化算法
优化算法
到目前为止,我们始终都是在使用梯度下降法学习,本文中,我们将使用一些更加高级的优化算法,利用这些优化算法,通常可以提高我们算法的收敛速度,并在最终得到更好的分离结果。这些方法可以加快学习速度,甚至可以为成本函数提供更好的最终值,在相同的结果下,有一个好的优化算法可以是等待几天和几个小时之间的差异。
我们想象一下成本函数 J J J,最小化损失函数就像找到丘陵的最低点,在训练的每一步中,都会按照某个方向更新参数,以尽可能达到最低点。它类似于最快的下山的路,如下图:

∂ J ∂ a = \frac{\partial J}{\partial a } = ∂a∂J=da
一、梯度下降算法
在机器学习中,最简单就是没有任何优化的梯度下降(GD,Gradient Descent),我们每一次循环都是对整个训练集 m m m进行学习,这叫做批量梯度下降(Batch Gradient Descent),我们之前说过了最核心的参数更新的公式,这里我们再来看一下:
for l = 1 , . . . , L l = 1, ..., L l=1,...,L:
W [ l ] = W [ l ] − α d W [ l ] W^{[l]} = W^{[l]} - \alpha \text{ } dW^{[l]} W[l]=W[l]−α dW[l]
b [ l ] = b [ l ] − α d b [ l ] b^{[l]} = b^{[l]} - \alpha \text{ } db^{[l]} b[l]=b[l]−α db[l]
- l l l是指当前的层数
- α \alpha α是学习率
所有的参数都被存储在一个parameters的字典类型中。们来看看它怎样实现的吧,虽然我们已经实现过它很多次了。
def update_parameters_with_gd(parameters, grads, learning_rate):
'''
使用梯度下降算法更新参数
:param parameters:包含需要更新的参数的python字典
parameters['W' + str(l)] = Wl
parameters['b' + str(l)] = bl
:param grads:python字典,用以更新参数的梯度值
grads['dW' + str(l)] = dWl
grads['db' + str(l)] = dbl
:param learning_rate:学习率
:return:
:parameters:更新之后的参数
'''
L = len(parameters) // 2
for l in range(L): # range(L):0 -> L-1
parameters["W" + str(l + 1)] = parameters["W" + str(l + 1)] - learning_rate * grads["dW" + str(l + 1)]
parameters["b" + str(l + 1)] = parameters["b" + str(l + 1)] - learning_rate * grads["db" + str(l + 1)]
return parameters
1.2 随机梯度下降算法
随机梯度下降算法(SGD,Stochastic Gradient Descent )是梯度下降算法一种变体,也相当于mini-batch size =1的mini-batch 梯度下降算法。随机梯度下降算法和梯度下降算法不同的是你一次只能在一个训练样本上计算梯度,而不是在整个训练集上计算梯度,下面来比较两者的不同之处:
- 梯度下降算法(Batch Gradient Descent)
#批量梯度下降,又叫梯度下降
X = data_input
Y = labels
parameters = initialize_parameters(layers_dims)
for i in range(0,num_iterations):
#前向传播
A,cache = forward_propagation(X,parameters)
#计算损失
cost = compute_cost(A,Y)
#反向传播
grads = backward_propagation(X,Y,cache)
#更新参数
parameters = update_parameters(parameters,grads)
- 随机梯度下降算法(Stochastic Gradient Descent)
#随机梯度下降算法:
X = data_input
Y = labels
parameters = initialize_parameters(layers_dims)
for i in (0,num_iterations):
for j in m:
#前向传播
A,cache = forward_propagation(X,parameters)
#计算成本
cost = compute_cost(A,Y)
#后向传播
grads = backward_propagation(X,Y,cache)
#更新参数
parameters = update_parameters(parameters,grads)
在随机梯度下降中,在更新梯度之前,只使用1个训练样本。 当训练集较大时,随机梯度下降可以更快,但是参数会向最小值摆动,而不是平稳地收敛,我们来看一下比较图:

"+"代表损失函数的最小值。随机梯度下降算法(SGD)在达到收敛时会有很多振荡。 但是,对于SGD而言,每个步骤的计算速度要快于GD,因为它仅使用一个训练示例(相对于GD的整个批次)。
1.3 mini-batch梯度下降算法
在实际中,更好的方法是使用小批量(mini-batch)梯度下降法,小批量梯度下降法是一种综合了梯度下降法和随机梯度下降法的方法,在它的每次迭代中,既不是选择全部的数据来学习,也不是选择一个样本来学习,而是把所有的数据集分割为一小块一小块的来学习,它会随机选择一小块(mini-batch),块大小一般为2的n次方倍。一方面,充分利用的GPU的并行性,更一方面,不会让计算时间特别长,来看一下比较图:
"+"代表损失函数的最小值。在优化算法中使用小批量通常可以加快优化速度。
我们要使用mini-batch要经过两个步骤:
- 把训练集打乱:但是X和Y依旧是一一对应的,之后,X的第 i i i列是与Y中的第 i i i个标签对应的样本。乱序步骤确保将样本被随机分成不同的小批次。如下图,X和Y的每一列代表一个样本:
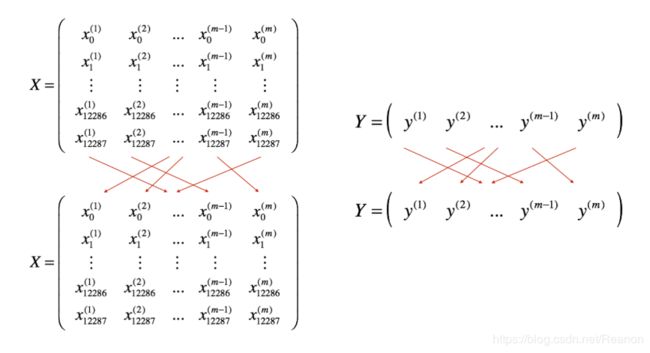
- .切分,我们把训练集打乱之后,我们就可以对它进行切分了。这里切分的大小
mini_batch_size=64,如下图:
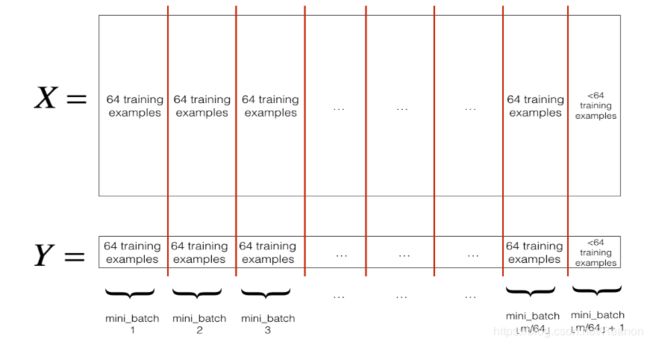 我们先来看看分割后如何获取第一第二个mini-batch吧。
我们先来看看分割后如何获取第一第二个mini-batch吧。
first_mini_batch_X = shuffled_X[:, 0 : mini_batch_size]
second_mini_batch_X = shuffled_X[:, mini_batch_size : 2 * mini_batch_size]
因为最后一个mini-batch 可能会小于mini_batch_size=64,所以我们这里用 ⌊ s ⌋ \lfloor s \rfloor ⌊s⌋ 来代表 s s s向下取整到最近的整数。在python中可以用math.floor(s)来实现。如果总体的样本数量不是mini_batch_size=64的整数倍,娜美就会有 ⌊ m m i n i _ b a t c h _ s i z e ⌋ \lfloor \frac{m}{mini\_batch\_size}\rfloor ⌊mini_batch_sizem⌋个包含64个样本的mini-bathes,而最后一个mini-batches大小为 m − m i n i _ b a t c h _ s i z e × ⌊ m m i n i _ b a t c h _ s i z e ⌋ m-mini_\_batch_\_size \times \lfloor \frac{m}{mini\_batch\_size}\rfloor m−mini_batch_size×⌊mini_batch_sizem⌋。
def random_mini_batches(X, Y, mini_batch_size=64, seed=0):
'''
从(X,Y)中创建一个随机的minibatches的列表。
:param X:输入数据
:param Y:真实标签值,1用蓝点、0用红点表示。形状是(1,样本数)
:param mini_batch_size:minibatches的大小
:param seed:随机种子
:return:
minibatches:一个同步列表(mini_batch_X,minibatch_Y.
'''
np.random.seed(seed)
m = X.shape[1]
mini_batches = []
# 第一步:打乱顺序
permutation = list(np.random.permutation(m)) # 它会返回一个长度为m的随机数组,且里面的数是0到m-1
shuffled_X = X[:, permutation] # 将每一列的数据按permutation的顺序来重新排列。
shuffled_Y = Y[:, permutation].reshape((1, m))
# 第二步:分割
num_complete_minibatches = math.floor(m / mini_batch_size) # 把你的训练集分割成多少份,请注意,如果值是99.99,那么返回值是99,剩下的0.99会被舍弃
for k in range(0, num_complete_minibatches):
mini_batch_X = shuffled_X[:, k * mini_batch_size:(k + 1) * mini_batch_size]
mini_batch_Y = shuffled_Y[:, k * mini_batch_size:(k + 1) * mini_batch_size]
mini_batch = (mini_batch_X, mini_batch_Y)
mini_batches.append(mini_batch)
# 如果训练集的大小刚好是mini_batch_size的整数倍,那么这里已经处理完了
# 如果训练集的大小不是mini_batch_size的整数倍,那么最后肯定会剩下一些,我们要把它处理了
if m % mini_batch_size != 0:
mini_batch_X = shuffled_X[:, mini_batch_size * num_complete_minibatches:]
mini_batch_Y = shuffled_Y[:, mini_batch_size * num_complete_minibatches:]
mini_batch = (mini_batch_X, mini_batch_Y) # 列表里包含着一个元组
mini_batches.append(mini_batch)
"""
#博主注:
#如果你不好理解的话请看一下下面的伪代码,看看X和Y是如何根据permutation来打乱顺序的。
x = np.array([[1,2,3,4,5,6,7,8,9],
[9,8,7,6,5,4,3,2,1]])
y = np.array([[1,0,1,0,1,0,1,0,1]])
random_mini_batches(x,y)
permutation= [7, 2, 1, 4, 8, 6, 3, 0, 5]
shuffled_X= [[8 3 2 5 9 7 4 1 6]
[2 7 8 5 1 3 6 9 4]]
shuffled_Y= [[0 1 0 1 1 1 0 1 0]]
"""
return mini_batches
二、动量梯度下降算法
由于mini-batch 梯度下降只看到了一个子集的参数更新,更新的方向有一定的差异,所以小批量梯度下降的路径将“振荡地”走向收敛,使用动量可以减少这些振荡,momentum梯度下降算法(Gradient descent with momentum)考虑了过去的梯度以平滑更新, 我们将把以前梯度的方向存储在变量 v v v中,从形式上讲,这将是前面的梯度的指数加权平均值(exponentially weighted average)。我们也可以把 v v v看作是滚下坡的速度,根据山坡的坡度建立动量。我们来看一下下面的图:
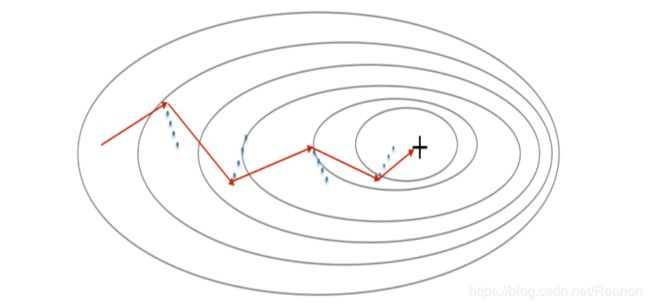
红色箭头显示具有动量的小批量梯度下降一步时所采取的方向,蓝色的点显示每个步骤的梯度方向(相对于当前的小批量),当然我们不仅要观察梯度,还要让 v v v影响梯度,然后朝 v v v方向前进一步,尽量让前进的方向指向最小值。
2.1 初始化 v v v
速度 v v v是一个需要用0初始化的python字典,它的关键字应当和grads保持一致:
for l = 1 , . . . , L l =1,...,L l=1,...,L:
v["dW" + str(l+1)] = ... #(numpy array of zeros with the same shape as parameters["W" + str(l+1)])
v["db" + str(l+1)] = ... #(numpy array of zeros with the same shape as parameters["b" + str(l+1)])
初始化velocity代码如下:
def initialize_velocity(parameters):
"""
初始化速度,velocity是一个字典:
- keys: "dW1", "db1", ..., "dWL", "dbL"
- values:与相应的梯度/参数维度相同的值为零的矩阵。
:param parameters:一个字典,包含了以下参数:
parameters["W" + str(l)] = Wl
parameters["b" + str(l)] = bl
返回:
v - 一个字典变量,包含了以下参数:
v["dW" + str(l)] = dWl的速度
v["db" + str(l)] = dbl的速度
"""
L = len(parameters) // 2
v = {}
for l in range(L):
v["dW" + str(l + 1)] = np.zeros_like(parameters["W" + str(l + 1)])
v["db" + str(l + 1)] = np.zeros_like(parameters["b" + str(l + 1)])
return v
2.2 使用momentum更新参数
momentum的更新规则如下:
for l = 1 , . . . , L l = 1, ..., L l=1,...,L:
{ v d W [ l ] = β v d W [ l ] + ( 1 − β ) d W [ l ] W [ l ] = W [ l ] − α v d W [ l ] \begin{cases} v_{dW^{[l]}} = \beta v_{dW^{[l]}} + (1 - \beta) dW^{[l]} \\ W^{[l]} = W^{[l]} - \alpha v_{dW^{[l]}} \end{cases} {vdW[l]=βvdW[l]+(1−β)dW[l]W[l]=W[l]−αvdW[l]
{ v d b [ l ] = β v d b [ l ] + ( 1 − β ) d b [ l ] b [ l ] = b [ l ] − α v d b [ l ] \begin{cases} v_{db^{[l]}} = \beta v_{db^{[l]}} + (1 - \beta) db^{[l]} \\ b^{[l]} = b^{[l]} - \alpha v_{db^{[l]}} \end{cases} {vdb[l]=βvdb[l]+(1−β)db[l]b[l]=b[l]−αvdb[l]
其中, L L L 是当前神经网络的层数, β \beta β是动量参数,一般取0.9, α \alpha α是学习率。所有的参数都被保存在parameters 字典中。
def update_parameters_with_momentum(parameters, grads, v, beta, learning_rate):
"""
使用动量更新参数
:param parameters:一个字典类型的变量,包含了以下字段:
parameters["W" + str(l)] = Wl
parameters["b" + str(l)] = bl
:param grads:个包含梯度值的字典变量,具有以下字段:
grads["dW" + str(l)] = dWl
grads["db" + str(l)] = dbl
:param v:包含当前速度的字典变量,具有以下字段:
v["dW" + str(l)] = ...
v["db" + str(l)] = ...
:param beta:超参数,动量,实数
:param learning_rate:学习率,实数
:return:
parameters 更新后的参数字典
v - 包含了更新后的速度变量
"""
L = len(parameters) // 2
for l in range(L):
# 计算速度(指数平均加权)
v["dW" + str(l + 1)] = beta * v["dW" + str(l + 1)] + (1 - beta) * grads["dW" + str(l + 1)]
v["db" + str(l + 1)] = beta * v["db" + str(l + 1)] + (1 - beta) * grads["db" + str(l + 1)]
# 更新参数
parameters["W" + str(l + 1)] = parameters["W" + str(l + 1)] - learning_rate * v["dW" + str(l + 1)]
parameters["b" + str(l + 1)] = parameters["b" + str(l + 1)] - learning_rate * v["db" + str(l + 1)]
return parameters, v
需要注意的是速度 v v v是用0来初始化的,因此,该算法需要经过几次迭代才能把速度提升上来并开始跨越更大步伐。当beta=0时,该算法相当于是没有使用momentum算法的标准的梯度下降算法。
动量 β β β越大,更新越平滑,因为我们越多地考虑过去的梯度。但是如果 β \beta β太大,它也可能使更新过于平滑。 β β β的一般取值范围从0.8到0.999。如果你不想调整这个,β=0.9通常是一个合理的默认值。
三、Adam优化算法
Adam(Adaptive moment Estimation)优化算法是用于训练神经网络的最有效的优化算法之一。它结合了RMSProp(Root mean square prop,均方根传播)和momentum优化算法。具体过程:
- 计算以前的梯度的指数加权平均值(exponentially weighted average),并将其存储在变量 v v v(偏差校正前)和 v c o r r e c t e d v^{corrected} vcorrected(偏差校正后)中。
- 计算以前梯度的平方的指数加权平均值(exponentially weighted average of the squares),并将其存储在变量 s s s(偏差校正前)和 s c o r r e c t e d s^{corrected} scorrected(偏差校正后)中。
- 将以上两种结合起来更新参数。
for l = 1 , . . . , L l = 1, ..., L l=1,...,L:
{ v d W [ l ] = β 1 v d W [ l ] + ( 1 − β 1 ) ∂ J ∂ W [ l ] v d W [ l ] c o r r e c t e d = v d W [ l ] 1 − ( β 1 ) t s d W [ l ] = β 2 s d W [ l ] + ( 1 − β 2 ) ( ∂ J ∂ W [ l ] ) 2 s d W [ l ] c o r r e c t e d = s d W [ l ] 1 − ( β 1 ) t W [ l ] = W [ l ] − α v d W [ l ] c o r r e c t e d s d W [ l ] c o r r e c t e d + ε \begin{cases} v_{dW^{[l]}} = \beta_1 v_{dW^{[l]}} + (1 - \beta_1) \frac{\partial \mathcal{J} }{ \partial W^{[l]} } \\ v^{corrected}_{dW^{[l]}} = \frac{v_{dW^{[l]}}}{1 - (\beta_1)^t} \\ s_{dW^{[l]}} = \beta_2 s_{dW^{[l]}} + (1 - \beta_2) (\frac{\partial \mathcal{J} }{\partial W^{[l]} })^2 \\ s^{corrected}_{dW^{[l]}} = \frac{s_{dW^{[l]}}}{1 - (\beta_1)^t} \\ W^{[l]} = W^{[l]} - \alpha \frac{v^{corrected}_{dW^{[l]}}}{\sqrt{s^{corrected}_{dW^{[l]}}} + \varepsilon} \end{cases} ⎩⎪⎪⎪⎪⎪⎪⎪⎪⎨⎪⎪⎪⎪⎪⎪⎪⎪⎧vdW[l]=β1vdW[l]+(1−β1)∂W[l]∂JvdW[l]corrected=1−(β1)tvdW[l]sdW[l]=β2sdW[l]+(1−β2)(∂W[l]∂J)2sdW[l]corrected=1−(β1)tsdW[l]W[l]=W[l]−αsdW[l]corrected+εvdW[l]corrected
其中:
- t :当前迭代的次数
- L 当前神经网络的层数
- β 1 \beta_1 β1和 β 2 \beta_2 β2 控制两个指数加权平均值的超参数averages.
- α \alpha α 学习率
- ε \varepsilon ε 一个非常小的数,用于避免除零操作,一般为 1 − 8 1^{-8} 1−8。
3.1 初始化Adam所需要的参数 v , s v, s v,s
它们需要被初始化为0,而且键值同grads一致。
for l = 1 , . . . , L l = 1, ..., L l=1,...,L:
v["dW" + str(l+1)] = ... #(numpy array of zeros with the same shape as parameters["W" + str(l+1)])
v["db" + str(l+1)] = ... #(numpy array of zeros with the same shape as parameters["b" + str(l+1)])
s["dW" + str(l+1)] = ... #(numpy array of zeros with the same shape as parameters["W" + str(l+1)])
s["db" + str(l+1)] = ... #(numpy array of zeros with the same shape as parameters["b" + str(l+1)])
初始化Adam优化算法的参数,代码如下:
def initialize_adam(parameters):
"""
将V和s都初始化为python字典
- keys: "dW1", "db1", ..., "dWL", "dbL"
- values:与对应的梯度/参数相同维度的值为零的numpy矩阵
:param parameters:包含了以下参数的字典变量:
parameters["W" + str(l)] = Wl
parameters["b" + str(l)] = bl
:return:
v - 包含梯度的指数加权平均值,字段如下:
v["dW" + str(l)] = ...
v["db" + str(l)] = ...
s - 包含平方梯度的指数加权平均值,字段如下:
s["dW" + str(l)] = ...
s["db" + str(l)] = ...
"""
L = len(parameters) // 2
v = {}
s = {}
# 初始化v,s.输入:parameters.输出:v,s.
for l in range(L):
v["dW" + str(l + 1)] = np.zeros_like(parameters["W" + str(l + 1)])
v["db" + str(l + 1)] = np.zeros_like(parameters["b" + str(l + 1)])
s["dW" + str(l + 1)] = np.zeros_like(parameters["W" + str(l + 1)])
s["db" + str(l + 1)] = np.zeros_like(parameters["b" + str(l + 1)])
return v, s
3.2 使用Adam优化算法更新参数
参数初始化完成了,我们就根据公式来更新参数:
{ v W [ l ] = β 1 v W [ l ] + ( 1 − β 1 ) ∂ J ∂ W [ l ] v W [ l ] c o r r e c t e d = v W [ l ] 1 − ( β 1 ) t s W [ l ] = β 2 s W [ l ] + ( 1 − β 2 ) ( ∂ J ∂ W [ l ] ) 2 s W [ l ] c o r r e c t e d = s W [ l ] 1 − ( β 2 ) t W [ l ] = W [ l ] − α v W [ l ] c o r r e c t e d s W [ l ] c o r r e c t e d + ε \begin{cases} v_{W^{[l]}} = \beta_1 v_{W^{[l]}} + (1 - \beta_1) \frac{\partial J }{ \partial W^{[l]} } \\ v^{corrected}_{W^{[l]}} = \frac{v_{W^{[l]}}}{1 - (\beta_1)^t} \\ s_{W^{[l]}} = \beta_2 s_{W^{[l]}} + (1 - \beta_2) (\frac{\partial J }{\partial W^{[l]} })^2 \\ s^{corrected}_{W^{[l]}} = \frac{s_{W^{[l]}}}{1 - (\beta_2)^t} \\ W^{[l]} = W^{[l]} - \alpha \frac{v^{corrected}_{W^{[l]}}}{\sqrt{s^{corrected}_{W^{[l]}}}+\varepsilon} \end{cases} ⎩⎪⎪⎪⎪⎪⎪⎪⎪⎨⎪⎪⎪⎪⎪⎪⎪⎪⎧vW[l]=β1vW[l]+(1−β1)∂W[l]∂JvW[l]corrected=1−(β1)tvW[l]sW[l]=β2sW[l]+(1−β2)(∂W[l]∂J)2sW[l]corrected=1−(β2)tsW[l]W[l]=W[l]−αsW[l]corrected+εvW[l]corrected
def update_parameters_with_adam(parameters, grads, v, s, t, learning_rate=0.01, beta1=0.9, beta2=0.999, epsilon=1e-8):
"""
使用Adam更新参数,,因为传输的字典,所以形参修改,实参也会被修改。
:param parameters: 包含了以下字段的字典:
parameters['W' + str(l)] = Wl
parameters['b' + str(l)] = bl
:param grads: 包含了梯度值的字典,有以下key值:
grads['dW' + str(l)] = dWl
grads['db' + str(l)] = dbl
:param v: Adam的变量,是一个字典类型的变量,包含梯度的指数加权平均值,字段如下:
v["dW" + str(l)] = ...
v["db" + str(l)] = ...
:param s: Adam的变量,是一个字典类型的变量,包含平方梯度的指数加权平均值,字段如下:
s["dW" + str(l)] = ...
s["db" + str(l)] = ...
:param t: 当前迭代的次数
:param learning_rate: 学习率
:param beta1: 动量,超参数,用于第一阶段,使得曲线的Y值不从0开始(参见天气数据的那个图)
:param beta2: RMSprop的一个参数,超参数
:param epsilon: 防止除零操作(分母为0)
:return:
parameters - 更新后的参数
v - 第一个梯度的移动平均值,是一个字典类型的变量
s - 平方梯度的移动平均值,是一个字典类型的变量
"""
L = len(parameters) // 2
v_corrected = {} # 偏差修正后的值
s_corrected = {} # 偏差修正后的值
for l in range(L):
# 计算momentum指数加权平均
v["dW" + str(l + 1)] = beta1 * v["dW" + str(l + 1)] + (1 - beta1) * grads["dW" + str(l + 1)]
v["db" + str(l + 1)] = beta1 * v["db" + str(l + 1)] + (1 - beta1) * grads["db" + str(l + 1)]
# 用RMsprop(均方根传播)计算平方梯度的移动平均值,输入:"s, grads , beta2",输出:"s"
s["dW" + str(l + 1)] = beta2 * s["dW" + str(l + 1)] + (1 - beta2) * np.power(grads["dW" + str(l + 1)], 2)
s["db" + str(l + 1)] = beta2 * s["db" + str(l + 1)] + (1 - beta2) * np.power(grads["db" + str(l + 1)], 2)
# 修正初期的偏差
v_corrected["dW" + str(l + 1)] = v["dW" + str(l + 1)] / (1 - np.power(beta1, t))
v_corrected["db" + str(l + 1)] = v["db" + str(l + 1)] / (1 - np.power(beta1, t))
s_corrected["dW" + str(l + 1)] = s["dW" + str(l + 1)] / (1 - np.power(beta1, t))
s_corrected["db" + str(l + 1)] = s["db" + str(l + 1)] / (1 - np.power(beta1, t))
# 更新参数:输入: "parameters, learning_rate, v_corrected, s_corrected, epsilon". 输出: "parameters".
parameters["W" + str(l + 1)] = parameters["W" + str(l + 1)] - learning_rate * (
v_corrected["dW" + str(l + 1)] / (np.sqrt(s_corrected["dW" + str(l + 1)]) + epsilon))
parameters["b" + str(l + 1)] = parameters["b" + str(l + 1)] - learning_rate * (
v_corrected["db" + str(l + 1)] / (np.sqrt(s_corrected["db" + str(l + 1)]) + epsilon))
return parameters, v, s
四、建立具体模型
我们之前已经实现过了一个三层的神经网络,我们将分别用它来测试我们的优化器的优化效果,我们先来看看我们的模型是什么样的:
def model(X, Y, layers_dims, optimizer, learning_rate=0.0007, mini_batch_size=64, beta=0.9, beta1=0.9, beta2=0.999,
epsilon=1e-8,
num_epochs=10000, print_cost=True, is_plot=True):
"""
可以运行在不同优化器模式下的3层神经网络模型。
:param X:输入数据,维度为(2,输入的数据集里面样本数量)
:param Y:与X对应的标签
:param layers_dims:包含层数和节点数量的列表
:param optimizer:字符串类型的参数,用于选择优化类型,【 "gd" | "momentum" | "adam" 】
:param learning_rate:学习率
mini_batch_size:每个小批量数据集的大小
:param beta:用于动量优化的一个超参数
:param beta1:用于计算平方梯度后的指数衰减的估计的超参数
:param beta2:用于计算梯度后的指数衰减的估计的超参数
:param epsilon:用于在Adam中避免除零操作的超参数,一般不更改
:param num_epochs:整个训练集的遍历次数,(视频2.9学习率衰减,1分55秒处,视频中称作“代”),相当于之前的num_iteration
:param print_cost:是否打印误差值,每遍历1000次数据集打印一次,但是每100次记录一个误差值,又称每1000代打印一次
:param is_plot:是否绘制出曲线图
:return:
parameters:包含了学习后的参数
"""
L = len(layers_dims)
costs = []
t = 0 # 每学习完一个minibatche就增加1
seed = 10 # 随机种子
parameters = initialize_parameters(layers_dims) # 初始化参数
# 选择优化器
if optimizer == "gd":
pass # 不使用任何优化器,直接使用梯度下降算法
elif optimizer == "momentum":
v = initialize_velocity(parameters) # 使用动量优化
elif optimizer == "adam":
v, s = initialize_adam(parameters) # 使用Adam优化
else:
print("optimizer参数错误")
exit(1)
# 开始学习
for i in range(num_epochs):
# 定义随机minibatches,我们每次遍历数据集之后增加种子以重新排列数据集,是每次数据的顺序不同
seed = seed + 1
minibatches = random_mini_batches(X, Y, mini_batch_size, seed)
for minibatch in minibatches:
# 选择一个minibatch
(minibatch_X, minibatch_Y) = minibatch
# 前向传播
A3, cache = forward_propagation(minibatch_X, parameters)
# 计算误差
cost = compute_cost(A3, minibatch_Y)
# 反向传播
grads = backward_propagation(minibatch_X, minibatch_Y, cache)
# 更新参数
if optimizer == "gd":
parameters = update_parameters_with_gd(parameters, grads, learning_rate)
elif optimizer == "momentum":
parameters, v = update_parameters_with_momentum(parameters, grads, v, beta, learning_rate)
elif optimizer == "adam":
t = t + 1 # Adam counter
parameters, v, s = update_parameters_with_adam(parameters, grads, v, s,
t, learning_rate, beta1, beta2, epsilon)
# 记录误差值
if i % 100 == 0:
costs.append(cost)
# 是否打印误差值
if print_cost and i % 1000 == 0:
print("第" + str(i) + "次遍历整个数据集,当前误差值:" + str(cost))
# 是否绘制曲线图
if is_plot:
plt.plot(costs)
plt.show()
plt.ylabel('cost')
plt.xlabel('epochs (per 100)')
plt.title("Learning rate = " + str(learning_rate))
plt.show()
return parameters
4.1 头文件
import numpy as np
import matplotlib.pyplot as plt
import scipy.io
import math
import sklearn
import sklearn.datasets
from opt_utils import load_params_and_grads, initialize_parameters, forward_propagation, backward_propagation
from opt_utils import compute_cost, predict, predict_dec, plot_decision_boundary, load_dataset
4.2 加载数据
train_X, train_Y = load_dataset()
layers_dims = [train_X.shape[0], 5, 2, 1]
4.3 使用三种不同的梯度下降算法优化
这里调用了三次,每次测试只需要调用其中一种即可
# # 1、使用普通的梯度下降算法
# parameters = model(train_X, train_Y, layers_dims, optimizer="gd")
# # 2、使用动量的梯度下降算法
# parameters = model(train_X, train_Y, layers_dims, beta=0.9, optimizer="momentum")
# 3、使用adam优化的梯度下降算法
parameters = model(train_X, train_Y, layers_dims, optimizer="adam")
这里有个待解决的问题是:在绘制costs的曲线图时前面的数据会堆叠在一起,暂时还没有找到具体的bug。
4.4 结果评价
# 进行预测
predictions = predict(train_X, train_Y, parameters)
# 绘制分类图
plt.rcParams['figure.figsize'] = (7.0, 4.0) # set default size of plots
plt.rcParams['image.interpolation'] = 'nearest'
plt.rcParams['image.cmap'] = 'gray'
# 绘制决策边界
plt.title("Model with Gradient Descent optimization")
axes = plt.gca()
axes.set_xlim([-1.5, 2.5])
axes.set_ylim([-1, 1.5])
plot_decision_boundary(lambda x: predict_dec(parameters, x.T), train_X, train_Y)
五、需要用到的组件
opt_utils.py
# -*- coding: utf-8 -*-
import numpy as np
import matplotlib.pyplot as plt
import sklearn
import sklearn.datasets
def sigmoid(x):
"""
Compute the sigmoid of x
Arguments:
x -- A scalar or numpy array of any size.
Return:
s -- sigmoid(x)
"""
s = 1 / (1 + np.exp(-x))
return s
def relu(x):
"""
Compute the relu of x
Arguments:
x -- A scalar or numpy array of any size.
Return:
s -- relu(x)
"""
s = np.maximum(0, x)
return s
def load_params_and_grads(seed=1):
np.random.seed(seed)
W1 = np.random.randn(2, 3)
b1 = np.random.randn(2, 1)
W2 = np.random.randn(3, 3)
b2 = np.random.randn(3, 1)
dW1 = np.random.randn(2, 3)
db1 = np.random.randn(2, 1)
dW2 = np.random.randn(3, 3)
db2 = np.random.randn(3, 1)
return W1, b1, W2, b2, dW1, db1, dW2, db2
def initialize_parameters(layer_dims):
"""
Arguments:
layer_dims -- python array (list) containing the dimensions of each layer in our network
Returns:
parameters -- python dictionary containing your parameters "W1", "b1", ..., "WL", "bL":
W1 -- weight matrix of shape (layer_dims[l], layer_dims[l-1])
b1 -- bias vector of shape (layer_dims[l], 1)
Wl -- weight matrix of shape (layer_dims[l-1], layer_dims[l])
bl -- bias vector of shape (1, layer_dims[l])
Tips:
- For example: the layer_dims for the "Planar Data classification model" would have been [2,2,1].
This means W1's shape was (2,2), b1 was (1,2), W2 was (2,1) and b2 was (1,1). Now you have to generalize it!
- In the for loop, use parameters['W' + str(l)] to access Wl, where l is the iterative integer.
"""
np.random.seed(3)
parameters = {}
L = len(layer_dims) # number of layers in the network
for l in range(1, L):
parameters['W' + str(l)] = np.random.randn(layer_dims[l], layer_dims[l - 1]) * np.sqrt(2 / layer_dims[l - 1])
parameters['b' + str(l)] = np.zeros((layer_dims[l], 1))
assert (parameters['W' + str(l)].shape == layer_dims[l], layer_dims[l - 1])
assert (parameters['W' + str(l)].shape == layer_dims[l], 1)
return parameters
def forward_propagation(X, parameters):
"""
Implements the forward propagation (and computes the loss) presented in Figure 2.
Arguments:
X -- input dataset, of shape (input size, number of examples)
parameters -- python dictionary containing your parameters "W1", "b1", "W2", "b2", "W3", "b3":
W1 -- weight matrix of shape ()
b1 -- bias vector of shape ()
W2 -- weight matrix of shape ()
b2 -- bias vector of shape ()
W3 -- weight matrix of shape ()
b3 -- bias vector of shape ()
Returns:
loss -- the loss function (vanilla logistic loss)
"""
# retrieve parameters
W1 = parameters["W1"]
b1 = parameters["b1"]
W2 = parameters["W2"]
b2 = parameters["b2"]
W3 = parameters["W3"]
b3 = parameters["b3"]
# LINEAR -> RELU -> LINEAR -> RELU -> LINEAR -> SIGMOID
z1 = np.dot(W1, X) + b1
a1 = relu(z1)
z2 = np.dot(W2, a1) + b2
a2 = relu(z2)
z3 = np.dot(W3, a2) + b3
a3 = sigmoid(z3)
cache = (z1, a1, W1, b1, z2, a2, W2, b2, z3, a3, W3, b3)
return a3, cache
def backward_propagation(X, Y, cache):
"""
Implement the backward propagation presented in figure 2.
Arguments:
X -- input dataset, of shape (input size, number of examples)
Y -- true "label" vector (containing 0 if cat, 1 if non-cat)
cache -- cache output from forward_propagation()
Returns:
gradients -- A dictionary with the gradients with respect to each parameter, activation and pre-activation variables
"""
m = X.shape[1]
(z1, a1, W1, b1, z2, a2, W2, b2, z3, a3, W3, b3) = cache
dz3 = 1. / m * (a3 - Y)
dW3 = np.dot(dz3, a2.T)
db3 = np.sum(dz3, axis=1, keepdims=True)
da2 = np.dot(W3.T, dz3)
dz2 = np.multiply(da2, np.int64(a2 > 0))
dW2 = np.dot(dz2, a1.T)
db2 = np.sum(dz2, axis=1, keepdims=True)
da1 = np.dot(W2.T, dz2)
dz1 = np.multiply(da1, np.int64(a1 > 0))
dW1 = np.dot(dz1, X.T)
db1 = np.sum(dz1, axis=1, keepdims=True)
gradients = {"dz3": dz3, "dW3": dW3, "db3": db3,
"da2": da2, "dz2": dz2, "dW2": dW2, "db2": db2,
"da1": da1, "dz1": dz1, "dW1": dW1, "db1": db1}
return gradients
def compute_cost(a3, Y):
"""
Implement the cost function
Arguments:
a3 -- post-activation, output of forward propagation
Y -- "true" labels vector, same shape as a3
Returns:
cost - value of the cost function
"""
m = Y.shape[1]
logprobs = np.multiply(-np.log(a3), Y) + np.multiply(-np.log(1 - a3), 1 - Y)
cost = 1. / m * np.sum(logprobs)
return cost
def predict(X, y, parameters):
"""
This function is used to predict the results of a n-layer neural network.
Arguments:
X -- data set of examples you would like to label
parameters -- parameters of the trained model
Returns:
p -- predictions for the given dataset X
"""
m = X.shape[1]
p = np.zeros((1, m), dtype=np.int)
# Forward propagation
a3, caches = forward_propagation(X, parameters)
# convert probas to 0/1 predictions
for i in range(0, a3.shape[1]):
if a3[0, i] > 0.5:
p[0, i] = 1
else:
p[0, i] = 0
# print results
# print ("predictions: " + str(p[0,:]))
# print ("true labels: " + str(y[0,:]))
print("Accuracy: " + str(np.mean((p[0, :] == y[0, :]))))
return p
def predict_dec(parameters, X):
"""
Used for plotting decision boundary.
Arguments:
parameters -- python dictionary containing your parameters
X -- input data of size (m, K)
Returns
predictions -- vector of predictions of our model (red: 0 / blue: 1)
"""
# Predict using forward propagation and a classification threshold of 0.5
a3, cache = forward_propagation(X, parameters)
predictions = (a3 > 0.5)
return predictions
def plot_decision_boundary(model, X, y):
# Set min and max values and give it some padding
x_min, x_max = X[0, :].min() - 1, X[0, :].max() + 1
y_min, y_max = X[1, :].min() - 1, X[1, :].max() + 1
h = 0.01
# Generate a grid of points with distance h between them
xx, yy = np.meshgrid(np.arange(x_min, x_max, h), np.arange(y_min, y_max, h))
# Predict the function value for the whole grid
Z = model(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
# Plot the contour and training examples
plt.contourf(xx, yy, Z, cmap=plt.cm.Spectral)
plt.ylabel('x2')
plt.xlabel('x1')
plt.scatter(X[0, :], X[1, :], c=np.squeeze(y), cmap=plt.cm.Spectral) # 这里需要将c=y 改成c=np.squeeze(y)
plt.show()
def load_dataset(is_plot=True):
np.random.seed(3)
train_X, train_Y = sklearn.datasets.make_moons(n_samples=300, noise=.2) # 300 #0.2
# Visualize the data
if is_plot:
plt.scatter(train_X[:, 0], train_X[:, 1], c=train_Y, s=40, cmap=plt.cm.Spectral);
train_X = train_X.T
train_Y = train_Y.reshape((1, train_Y.shape[0]))
return train_X, train_Y