机器学习:SVM作业编程实现(一)
SVM作业编程实现(一)
本次作业是实现SVM学习方法中序列最优化算法(SMO),其中my_svm.m和my_svmtrain.m与此次作业相关的两个文件。
my_svm.m
%Load the sample data, which includes Fisher's iris data of 5 measurements on a sample of 150 irises.
load fisheriris
%Create data, a two-column matrix containing sepal length and sepal width measurements for 150 irises.
data = [meas(:,1), meas(:,2)];
% From the species vector, create a new column vector, groups, to classify data into two groups: Setosa and non-Setosa.
groups = ismember(species,'setosa');
%Randomly select training and test sets.
[train, test] = crossvalind('holdOut',groups);
cp = classperf(groups);
%Train an SVM classifier using a linear kernel function and plot the grouped data.
svmStruct = my_svmtrain(data(train,:),groups(train),'showplot',true,'METHOD','SMO');
% Add a title to the plot
title(sprintf('Kernel Function: %s',...
func2str(svmStruct.KernelFunction)),...
'interpreter','none');
% Classify the test set using svmclassify
% See how well the classifier performed
classperf(cp,classes,test);
cp.CorrectRateLIA = ismember(A,B) 返回的是和A一样大小的数组,判断A中对应的元素在B中是否存在,若存在则返回1,否则为0.
[TRAIN,TEST] = crossvalind(‘HoldOut’,N,P)为交叉验证函数,‘HoldOut’方法从N个观测中随机分出P*N个作为验证数据,其中P位于(0,1),缺省值为0.5.TRAIN和TEST返回的向量是由0或1构成,代表对应数据是否是TRAIN或者TEST数据。这里用到的是[TRAIN,TEST] = crossvalind(‘HoldOut’,GROUPS),是对GROUPS进行选取到训练和测试中。
CP = classperf(GROUNDTRUTH)创建一个新的分类验证对象,GROUNDTRUTH是观测对象的真实分类。
my_svmtrain.m
需要将my_svmtrain.m中的自带函数seqminopt用自己编写的函数替换。
function [svm_struct, svIndex] = my_svmtrain(training, groupnames, varargin)
%SVMTRAIN trains a support vector machine classifier
%
% SVMStruct = SVMTRAIN(TRAINING,GROUP) trains a support vector machine
% classifier using data TRAINING taken from two groups given by GROUP.
% SVMStruct contains information about the trained classifier, including
% the support vectors, that is used by SVMCLASSIFY for classification.
% GROUP is a column vector of values of the same length as TRAINING that
% defines two groups. Each element of GROUP specifies the group the
% corresponding row of TRAINING belongs to. GROUP can be a numeric
% vector, a string array, or a cell array of strings. SVMTRAIN treats
% NaNs or empty strings in GROUP as missing values and ignores the
% corresponding rows of TRAINING.
%
% SVMTRAIN(...,'KERNEL_FUNCTION',KFUN) allows you to specify the kernel
% function KFUN used to map the training data into kernel space. The
% default kernel function is the dot product. KFUN can be one of the
% following strings or a function handle:
%
% 'linear' Linear kernel or dot product
% 'quadratic' Quadratic kernel
% 'polynomial' Polynomial kernel (default order 3)
% 'rbf' Gaussian Radial Basis Function kernel
% 'mlp' Multilayer Perceptron kernel (default scale 1)
% function A kernel function specified using @,
% for example @KFUN, or an anonymous function
%
% A kernel function must be of the form
%
% function K = KFUN(U, V)
%
% The returned value, K, is a matrix of size M-by-N, where U and V have M
% and N rows respectively. If KFUN is parameterized, you can use
% anonymous functions to capture the problem-dependent parameters. For
% example, suppose that your kernel function is
%
% function k = kfun(u,v,p1,p2)
% k = tanh(p1*(u*v')+p2);
%
% You can set values for p1 and p2 and then use an anonymous function:
% @(u,v) kfun(u,v,p1,p2).
%
% SVMTRAIN(...,'RBF_SIGMA',SIGMA) allows you to specify the scaling
% factor, sigma, in the radial basis function kernel.
%
% SVMTRAIN(...,'POLYORDER',ORDER) allows you to specify the order of a
% polynomial kernel. The default order is 3.
%
% SVMTRAIN(...,'MLP_PARAMS',[P1 P2]) allows you to specify the
% parameters of the Multilayer Perceptron (mlp) kernel. The mlp kernel
% requires two parameters, P1 and P2, where K = tanh(P1*U*V' + P2) and P1
% > 0 and P2 < 0. Default values are P1 = 1 and P2 = -1.
%
% SVMTRAIN(...,'METHOD',METHOD) allows you to specify the method used
% to find the separating hyperplane. Options are
%
% 'QP' Use quadratic programming (requires the Optimization Toolbox)
% 'SMO' Use Sequential Minimal Optimization method
% 'LS' Use least-squares method
%
% If you have the Optimization Toolbox, then the QP method is the default
% method. If not, the default method is SMO. When using the QP method,
% the classifier is a 2-norm soft-margin support vector machine.
%
% SVMTRAIN(...,'QUADPROG_OPTS',OPTIONS) allows you to pass an OPTIONS
% structure created using OPTIMSET to the QUADPROG function when using
% the 'QP' method. See help optimset for more details.
%
% SVMTRAIN(...,'SMO_OPTS',SMO_OPTIONS) allows you to set options for the
% 'SMO' method. SMO_OPTIONS should be created using the function
% SVMSMOSET.
%
% SVMTRAIN(...,'BOXCONSTRAINT',C) allows you to set the box constraint C
% for the soft margin. The default value is 1. C can be a scalar or a
% vector of the same length as the training data. Note that in older
% versions of Bioinformatics Toolbox the default value for C was
% 1/sqrt(eps) which will only classify separable data.
%
% SVMTRAIN(...,'AUTOSCALE', AUTOSCALEVAL) allows you to specify whether
% or not to automatically shift and scale the data points before
% training. Default is true.
%
% SVMTRAIN(...,'SHOWPLOT',true), when used with two-dimensional data,
% creates a plot of the grouped data and plots the separating line for
% the classifier.
%
% Example:
% % Load the data and select features for classification
% load fisheriris
% data = [meas(:,1), meas(:,2)];
% % Extract the Setosa class
% groups = ismember(species,'setosa');
% % Randomly select training and test sets
% [train, test] = crossvalind('holdOut',groups);
% cp = classperf(groups);
% % Use a linear support vector machine classifier
% svmStruct = svmtrain(data(train,:),groups(train),'showplot',true);
% % Add a title to the plot
% title(sprintf('Kernel Function: %s',...
% func2str(svmStruct.KernelFunction)),...
% 'interpreter','none');
% % Classify the test set using svmclassify
% classes = svmclassify(svmStruct,data(test,:),'showplot',true);
% % See how well the classifier performed
% classperf(cp,classes,test);
% cp.CorrectRate
%
% See also CLASSIFY, CLASSPERF, CROSSVALIND, KNNCLASSIFY, QUADPROG,
% SVMCLASSIFY, SVMSMOSET.
% Copyright 2004-2008 The MathWorks, Inc.
% $Revision: 1.1.12.13 $ $Date: 2008/06/16 16:32:47 $
% References:
%
% [1] Cristianini, N., Shawe-Taylor, J An Introduction to Support
% Vector Machines, Cambridge University Press, Cambridge, UK. 2000.
% http://www.support-vector.net
% [2] Kecman, V, Learning and Soft Computing,
% MIT Press, Cambridge, MA. 2001.
% [3] Suykens, J.A.K., Van Gestel, T., De Brabanter, J., De Moor, B.,
% Vandewalle, J., Least Squares Support Vector Machines,
% World Scientific, Singapore, 2002.
% [4] J.C. Platt: A Fast Algorithm for Training Support Vector
% Machines, Advances in Kernel Methods - Support Vector Learning,
% B. Sch?lkopf, C. Burges, and A. Smola, eds., MIT Press, 1998.
% [5] J.C. Platt: Fast Training of Support Vector Machines using
% Sequential Minimal Optimization Microsoft Research Technical
% Report MSR-TR-98-14, 1998.
% [6] http://www.kernel-machines.org/papers/tr-30-1998.ps.gz
%
% SVMTRAIN(...,'KFUNARGS',ARGS) allows you to pass additional
% arguments to kernel functions.
% check inputs
bioinfochecknargin(nargin,2,mfilename)
% set defaults
plotflag = false;
% The large scale solver cannot handle this type of problem, so turn it
% off.
qp_opts = optimset('LargeScale','Off','display','off');
smo_opts = svmsmoset;
kfunargs = {};
setPoly = false; usePoly = false;
setMLP = false; useMLP = false;
setSigma = false; useSigma = false;
autoScale = true;
% default optimization method
if ~isempty(which('quadprog'))
optimMethod = 'QP';
else
optimMethod = 'SMO';
end
% set default kernel function
kfun = @linear_kernel;
numoptargs = nargin -2;
optargs = varargin;
% check group is a vector -- though char input is special...
if ~isvector(groupnames) && ~ischar(groupnames)
error('Bioinfo:svmtrain:GroupNotVector',...
'Group must be a vector.');
end
% grp2idx sorts a numeric grouping var ascending, and a string grouping
% var by order of first occurrence
[groupIndex, groupString] = grp2idx(groupnames);
% make sure that the data are correctly oriented.
if size(groupnames,1) == 1
groupnames = groupnames';
end
% make sure data is the right size
if size(training,1) ~= length(groupnames)
if size(training,2) == length(groupnames)
training = training';
else
error('Bioinfo:svmtrain:DataGroupSizeMismatch',...
'GROUP and TRAINING must have the same number of rows.')
end
end
% check for NaN in data matrix:
if any(isnan(training(:)))
error('Bioinfo:svmtrain:NaNinDataMatrix', ...
'TRAINING data must not contain missing values');
end
% NaNs are treated as unknown classes and are removed from the training
% data
nans = isnan(groupIndex);
if any(nans)
training(nans,:) = [];
groupIndex(nans) = [];
end
ngroups = length(groupString);
nPoints = length(groupIndex);
% set default value of box constraint
boxconstraint = ones(nPoints, 1);
% convert to 1, -1.
groupIndex = 1 - (2* (groupIndex-1));
% handle optional arguments
if numoptargs >= 1
if rem(numoptargs,2)== 1
error('Bioinfo:svmtrain:IncorrectNumberOfArguments',...
'Incorrect number of arguments to %s.',mfilename);
end
okargs = {'kernel_function','method','showplot','kfunargs',...
'quadprog_opts','polyorder','mlp_params',...
'boxconstraint','rbf_sigma','autoscale', 'smo_opts'};
end
%选定优化方法
optimMethod= 'SMO';
plotflag= 'true';
% plot the data if requested
if plotflag
[hAxis,hLines] = svmplotdata(training,groupIndex);
legend(hLines,cellstr(groupString));
end
% autoscale data if required, we can not use the zscore function here,
% because we need the shift and scale values.
scaleData = [];
if autoScale
scaleData.shift = - mean(training);
stdVals = std(training);
scaleData.scaleFactor = 1./stdVals;
% leave zero-variance data unscaled:
scaleData.scaleFactor(~isfinite(scaleData.scaleFactor)) = 1;
% shift and scale columns of data matrix:
for c = 1:size(training, 2)
training(:,c) = scaleData.scaleFactor(c) * ...
(training(:,c) + scaleData.shift(c));
end
end
% if we have a kernel that takes extra arguments we must define a new
% kernel function handle to be passed to seqminopt
if ~isempty(kfunargs)
tmp_kfun = @(x,y) feval(kfun, x,y, kfunargs{:});
else
tmp_kfun = kfun;
end
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%请使用自己设计的SMO算法替代原Matlab的SMO算法
[alpha bias] = seqminopt(training, groupIndex, ...
boxconstraint, tmp_kfun, smo_opts);
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
svIndex = find(alpha > sqrt(eps));
sv = training(svIndex,:);
alphaHat = groupIndex(svIndex).*alpha(svIndex);
svm_struct.SupportVectors = sv;
svm_struct.Alpha = alphaHat;
svm_struct.Bias = bias;
svm_struct.KernelFunction = kfun;
svm_struct.KernelFunctionArgs = kfunargs;
svm_struct.GroupNames = groupnames;
svm_struct.SupportVectorIndices = svIndex;
svm_struct.ScaleData = scaleData;
svm_struct.FigureHandles = [];
if plotflag
hSV = svmplotsvs(hAxis,hLines,groupString,svm_struct);
svm_struct.FigureHandles = {hAxis,hLines,hSV};
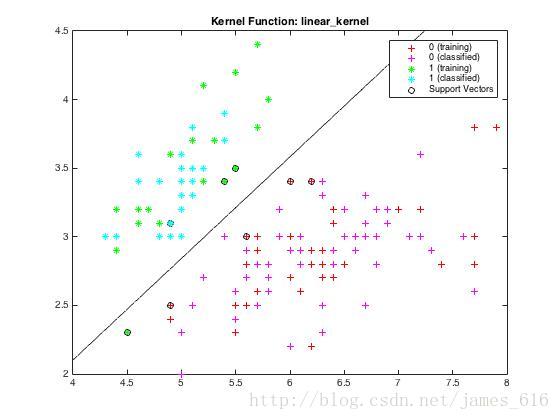
end运行my_svm.m可以得到下图结果,由训练样本得到分类面,并对测试样本进行分类。
下面开始解读my_svmtrain.m
首先是函数的定义
function [svm_struct, svIndex] = my_svmtrain(training, groupnames, varargin)
在这里需要理解一下varargin,在matlab中, varargin提供了一种函数可变参数列表机制。 就是说, 使用了“可变参数列表机制”的函数允许调用者调用该函数时根据需要来改变输入参数的个数。
注释部分是对参数的一些说明,包括核函数的选择、优化方法的选择以及二维数据的可视化选项。如果不对核函数特别要求,则默认内积。如果不对优化方法设定,则默认用 quadratic programming方法,在我们的作业中则是要用SMO的优化方法。
程序正式部分开始是对输入数据的一些检查,暂且可以不考虑。直接看
[groupIndex, groupString] = grp2idx(groupnames);
函数grp2idx能够返回向量groupIndex由1开始到k的整数,代表不同的类别以及向量groupString代表具体的类别名。对于我们的二分类问题则是groupIndex为1和2,groupString对应‘1’和‘0’。
接下来的部分还是对数据的一些检查,直接看
groupIndex = 1 - (2* (groupIndex-1));
因为groupIndex为1和2,通过这一步就可以变成1和-1,即为SVM中标准类别。
优化方法选为SMO,接下来是对数据大小进行尺度变换的操作,也可以不用考虑,直奔主题替换以下代码。
[alpha bias] = seqminopt(training, groupIndex, ...
boxconstraint, tmp_kfun, smo_opts);这一部分将在下一篇博客中具体介绍,欢迎继续浏览!