Unet分割直肠肿瘤图像
最近做了一个关于分割直肠肿瘤的项目,用的是Unet模型,从一开始就遇到了许许多多的问题,比如读dcm图像的问题,训练结果全黑或全灰的问题,但功夫不负有心人,经过几个星期的调试,最终终于得到了还不错的结果。下面想给大家分享一下我的源码,并说明一下相关问题的解决方法。
我会分块对代码进行分析,为了大家调试方便,只需要将我的代码块从上到下连起来就可以正常运行了。
代码和数据集都在github和网盘上:https://github.com/zf617707527/Unet1.git
网盘:链接:https://pan.baidu.com/s/1ZtK6MEPozLC7K8BnSO9ncg
提取码:u4wg
所依赖的库
import cv2
from keras.preprocessing.image import ImageDataGenerator
import os
import numpy as np
from keras.models import Model
from keras.layers import Input, concatenate, Conv2D, MaxPooling2D, Conv2DTranspose, Cropping2D
from keras.optimizers import Adam
from keras.callbacks import ModelCheckpoint
from keras import backend as K
from skimage import io
import h5py
import SimpleITK as sitk
装库的时候也遇到了一些问题,我一般习惯于把pycharm和anaconda结合起来用,将anaconda的库配置到pycharm中,就可以省去很多装库的问题。但是有一些库还是很难装,经常安装失败,大家可以试一试后再用anaconda的命令行进行安装,我用这个方法还是挺有效的。
所有的函数块
切片函数
def getRangImageDepth(image):
firstflag = True
startposition = 0
endposition = 0
for z in range(image.shape[0]):
notzeroflag = np.max(image[z])
if notzeroflag and firstflag:
startposition = z
firstflag = False
if notzeroflag:
endposition = z
return startposition, endposition
这个函数进行了切片操作,只对有肿瘤的CT图和相应的掩模图进行训练,减少其它无肿瘤图像对训练模型的影响。
读h5文件
class HDF5DatasetGenerator:
def __init__(self, dbPath, batchSize, preprocessors=None,
aug=None, binarize=True, classes=2):
self.batchSize = batchSize
self.preprocessors = preprocessors
self.aug = aug
self.binarize = binarize
self.classes = classes
self.db = h5py.File(dbPath)
self.numImages = self.db["images"].shape[0]
#print('&&&&&')
#print(self.db["images"].shape[0])
#print(self.db["images"].shape)
# self.numImages = total
print("total images:", self.numImages)
self.num_batches_per_epoch = int((self.numImages - 1) / batchSize) + 1
def generator(self, shuffle=True, passes=np.inf):
epochs = 0 #原来的值为0
while epochs < passes:
shuffle_indices = np.arange(self.numImages)
shuffle_indices = np.random.permutation(shuffle_indices)
for batch_num in range(self.num_batches_per_epoch):
start_index = batch_num * self.batchSize
end_index = min((batch_num + 1) * self.batchSize, self.numImages)
# h5py get item by index,参数为list,而且必须是增序
batch_indices = sorted(list(shuffle_indices[start_index:end_index]))
#print('******')
#print(batch_indices)
images = self.db["images"][batch_indices, :, :, :]
labels = self.db["masks"][batch_indices, :, :, :]
# if self.binarize:
# labels = np_utils.to_categorical(labels, self.classes)
if self.preprocessors is not None:
procImages = []
for image in images:
for p in self.preprocessors:
image = p.preprocess(image)
procImages.append(image)
images = np.array(procImages)
if self.aug is not None:
# 不知道意义何在?本身images就有batchsize个了
(images, labels) = next(self.aug.flow(images, labels,
batch_size=self.batchSize))
yield (images, labels)
epochs += 1
def close(self):
self.db.close()
用来读h5文件的数据
写h5文件
class HDF5DatasetWriter:
def __init__(self, image_dims, mask_dims, outputPath, bufSize=200):
"""
Args:
- bufSize: 当内存储存了bufSize个数据时,就需要flush到外存
"""
if os.path.exists(outputPath):
raise ValueError("The supplied 'outputPath' already"
"exists and cannot be overwritten. Manually delete"
"the file before continuing", outputPath)
self.db = h5py.File(outputPath, "w")
self.data = self.db.create_dataset("images", image_dims, dtype="float")
self.masks = self.db.create_dataset("masks", mask_dims, dtype="int")
self.bufSize = bufSize
self.buffer = {"data": [], "masks": []}
self.idx = 0
def add(self, rows, masks):
# extend() 函数用于在列表末尾一次性追加另一个序列中的多个值(用新列表扩展原来的列表)
# 注意,用extend还有好处,添加的数据不会是之前list的引用!!
self.buffer["data"].extend(rows)
self.buffer["masks"].extend(masks)
print("len ", len(self.buffer["data"]))
if len(self.buffer["data"]) >= self.bufSize:
self.flush()
def flush(self):
i = self.idx + len(self.buffer["data"])
self.data[self.idx:i, :, :, :] = self.buffer["data"]
self.masks[self.idx:i, :, :, :] = self.buffer["masks"]
print("h5py have writen %d data" % i)
self.idx = i
self.buffer = {"data": [], "masks": []}
def close(self):
if len(self.buffer["data"]) > 0:
self.flush()
self.db.close()
return self.idx
用来写数据到h5文件
得到CT的hu值函数
def get_pixels_hu(scans):
image = np.stack([ sitk.GetArrayFromImage(s)[0] for s in scans])
return np.array(image, dtype=np.int16)
因为我们的CT图是用SimpleITK读的,所读出来的就是hu值,再这里只做了将读取的CT图聚合到一起的操作。
窗口函数
def transform_ctdata(self, windowWidth, windowCenter, normal=False):
"""
注意,这个函数的self.image一定得是float类型的,否则就无效!
return: trucated image according to window center and window width
"""
minWindow = float(windowCenter) - 0.5 * float(windowWidth)
print('<<<')
print(self)
newimg = (self - minWindow) / float(windowWidth)
newimg[newimg < 0] = 0
newimg[newimg > 1] = 0
if not normal:
newimg = (newimg * 255).astype('uint8')
return newimg
这个函数是做了一个窗口化的操作,为了增强对比度,具体原理就是设置一个hu值的窗口,我们是统计直肠肿瘤的hu值大概确定了直肠肿瘤所处的hu值范围,然后将这个范围做成窗口,窗口内的也就是目标区域不变,其他区域都变成白色,使目标区域与周围其他组织对比度更大,增强学习的效果。选择一个合适的窗口是很关键的,如果窗口选的好,最后就算输入一个不含目标区域的图像,经过窗口化处理就会变成一个全白的图像,这样模型就同时具有了识别不含肿瘤CT的功能
直方图均衡化
def clahe_equalized(imgs,start,end):
assert (len(imgs.shape)==3) #3D arrays assert用于声明该条件是对的,也即输入的图片是三通道的,如果不是会有一个报错!
#create a CLAHE object (Arguments are optional).
clahe = cv2.createCLAHE(clipLimit=2.0, tileGridSize=(8,8))#自适应的均衡化过程,这些参数都是固定的。
imgs_equalized = np.empty(imgs.shape)
for i in range(start, end+1):
imgs_equalized[i,:,:] = clahe.apply(np.array(imgs[i,:,:], dtype = np.uint8))
return imgs_equalized
主程序
数据增强
TOTAL = 600#训练集数目
VAL_TOTAL = 35#测试集数目
seed=42
data_gen_args = dict(rotation_range=3,
width_shift_range=0.01,
height_shift_range=0.01,
shear_range=0.01,
zoom_range=0.01,
fill_mode='nearest')
image_datagen = ImageDataGenerator(**data_gen_args)
mask_datagen = ImageDataGenerator(**data_gen_args)
其中的TOTAL是很重要的参数,这里的训练集数目要自己数出来,这里TOTAL还会影响到steps_per_epoch这个参数,因为这个参数设置的为TOTAL/BATCH_SIZE,决定了每一个epoch将训练集分成了多少份进行训练。但是经过我试验TOTAL并不一定要是训练集的数目,不一样也不会报错,而且会影响训练的效果,之前这个TOTAL设的不对导致结果总是全黑或全灰,而我把改到600时效果就很不错,所以这个参数是很重要的。
训练数据提取和数据预处理
dataset = HDF5DatasetWriter(image_dims=(TOTAL, 512, 512, 1),
mask_dims=(TOTAL, 512, 512, 1),
outputPath="train_liver.h5") # 保存到文件中去
for i in range(1, 18): # 前18个人作为训练集
full_images = [] # 后面用来存储目标切片的列表
full_livers = [] # 功能同上
# 注意不同的系统,文件分割符的区别
label_path = 'data1/%d/mask' %i #读取掩模图片
data_path = 'data1/%d/arterial phase' %i #加入CT图片的位置
liver_slices = [cv2.cvtColor((cv2.imread(label_path + '/' + s)),cv2.COLOR_BGR2GRAY)for s in os.listdir(label_path)]
livers = np.stack([s for s in liver_slices])#所有的直肠的图片的矩阵聚合在一起!
#print(data_path + '/' + os.listdir(data_path)[0])
image_slices = [sitk.ReadImage(data_path + '/' + s) for s in os.listdir(data_path)]
images = get_pixels_hu(image_slices)#对切片进行CT-HU值还原!
images = transform_ctdata(images,250, 75)#增强对比度
start, end = getRangImageDepth(livers)#得到有肿瘤的图像
# print('(((')
# print(start,end)
images = clahe_equalized(images, start, end) #对所有的图片都进行了图片直方图的均衡化!
images /= 255#像素值归为【0,1】,这样就利于1后续生成掩模图了
total = end - start + 1#经过切片后图片的总数
print("%d person, total slices %d" % (i, total))
# 首和尾目标区域都太小,舍弃
images = images[start:end]
print("%d person, images.shape:(%d,)" % (i, images.shape[0]))
livers[np.array(livers) > 0] = 1#对掩模图进行了归一化处理
livers = livers[start:end]#为了与images匹配,所以进行了相同的切片操作!
full_images.append(images)
full_livers.append(livers)
full_images = np.vstack(full_images) #也是叠加矩阵的作用
full_images = np.expand_dims(full_images, axis=-1) #expand_dims就是对矩阵的形状进行调制,axis=-1,表示对其进行了最后面的插入。
full_livers = np.vstack(full_livers)
full_livers = np.expand_dims(full_livers, axis=-1)
# 数据增强!
image_datagen.fit(full_images, augment=True, seed=seed)
mask_datagen.fit(full_livers, augment=True, seed=seed)
image_generator = image_datagen.flow(full_images, seed=seed)
mask_generator = mask_datagen.flow(full_livers, seed=seed)
train_generator = zip(image_generator, mask_generator) # 将两个图片集进行了叠加
x = []
y = []
i = 0
for x_batch, y_batch in train_generator:
i += 1
x.append(x_batch)
y.append(y_batch)
if i >= 2: # 因为我不需要太多的数据
break
x = np.vstack(x)
y = np.vstack(y)
dataset.add(full_images, full_livers)
dataset.add(x, y)
dataset.close()
测试数据提取和数据预处理
full_images2 = []
full_livers2 = []
for i in range(105,108):# 后3个人作为测试样本
label_path = 'data1/%d/mask' %i #读取掩模图片
data_path = 'data1/%d/arterial phase' %i #加入CT图片的位置
liver_slices = [cv2.cvtColor((cv2.imread(label_path + '/' + s)),cv2.COLOR_BGR2GRAY)for s in os.listdir(label_path)]
#listdir需要接受到的是一个绝对路径//它返回该路径下的所有文件及其目录
# s.pixel_array 获取dicom格式中的像素值
livers = np.stack([s for s in liver_slices])#所有的肝部的图片的矩阵聚合在一起!
image_slices = [sitk.ReadImage(data_path+'/'+s) for s in os.listdir(data_path)]
images = get_pixels_hu(image_slices) # 对切片进行CT-HU值还原!
images = transform_ctdata(images, 250, 75)
start, end = getRangImageDepth(livers)
# print(')))')
# print(start,end)
images = clahe_equalized(images, start, end) # 对所有的图片都进行了图片直方图的均衡化!
images /= 255 # 像素值归为【0,1】,这样就利于1后续生成掩模图了
images = images[start:end]
print("%d person, images.shape:(%d,)" % (i, images.shape[0]))
livers[livers > 0] = 1
livers = livers[start:end] # 为了与images匹配,所以进行了相同的切片操作!
full_images2.append(images)
full_livers2.append(livers)
full_images2 = np.vstack(full_images2)
full_images2 = np.expand_dims(full_images2, axis=-1)
full_livers2 = np.vstack(full_livers2)
full_livers2 = np.expand_dims(full_livers2, axis=-1)
dataset = HDF5DatasetWriter(image_dims=(full_images2.shape[0], full_images2.shape[1], full_images2.shape[2], 1),
mask_dims=(full_images2.shape[0], full_images2.shape[1], full_images2.shape[2], 1),
outputPath="val_liver.h5")#这里的路径其实就是val_outputPath
dataset.add(full_images2, full_livers2)
print("total images in val ", dataset.close())
K.set_image_data_format('channels_last')
Unet模型的构建
def dice_coef(y_true, y_pred):
smooth = 1.
y_true_f = K.flatten(y_true)
y_pred_f = K.flatten(y_pred)
intersection = K.sum(y_true_f * y_pred_f)
return (2. * intersection + smooth) / (K.sum(y_true_f) + K.sum(y_pred_f) + smooth)
def dice_coef_loss(y_true, y_pred):
return -dice_coef(y_true, y_pred)
def get_crop_shape(target, refer):
# width, the 3rd dimension
print(target.shape)
print(refer._keras_shape)
cw = (target._keras_shape[2] - refer._keras_shape[2])
assert (cw >= 0)
if cw % 2 != 0:
cw1, cw2 = int(cw / 2), int(cw / 2) + 1
else:
cw1, cw2 = int(cw / 2), int(cw / 2)
# height, the 2nd dimension
ch = (target._keras_shape[1] - refer._keras_shape[1])
assert (ch >= 0)
if ch % 2 != 0:
ch1, ch2 = int(ch / 2), int(ch / 2) + 1
else:
ch1, ch2 = int(ch / 2), int(ch / 2)
return (ch1, ch2), (cw1, cw2)
def get_unet():
inputs = Input((IMG_HEIGHT, IMG_WIDTH, 1))
conv1 = Conv2D(32, (3, 3), activation='relu', padding='same')(inputs)
conv1 = Conv2D(32, (3, 3), activation='relu', padding='same')(conv1)
pool1 = MaxPooling2D(pool_size=(2, 2))(conv1)
conv2 = Conv2D(64, (3, 3), activation='relu', padding='same')(pool1)
conv2 = Conv2D(64, (3, 3), activation='relu', padding='same')(conv2)
pool2 = MaxPooling2D(pool_size=(2, 2))(conv2)
conv3 = Conv2D(128, (3, 3), activation='relu', padding='same')(pool2)
conv3 = Conv2D(128, (3, 3), activation='relu', padding='same')(conv3)
pool3 = MaxPooling2D(pool_size=(2, 2))(conv3)
conv4 = Conv2D(256, (3, 3), activation='relu', padding='same')(pool3)
conv4 = Conv2D(256, (3, 3), activation='relu', padding='same')(conv4)
pool4 = MaxPooling2D(pool_size=(2, 2))(conv4)
conv5 = Conv2D(512, (3, 3), activation='relu', padding='same')(pool4)
conv5 = Conv2D(512, (3, 3), activation='relu', padding='same')(conv5)
up_conv5 = Conv2DTranspose(256, (2, 2), strides=(2, 2), padding='same')(conv5)
ch, cw = get_crop_shape(conv4, up_conv5)
crop_conv4 = Cropping2D(cropping=(ch, cw), data_format="channels_last")(conv4)
up6 = concatenate([up_conv5, crop_conv4], axis=3)
conv6 = Conv2D(256, (3, 3), activation='relu', padding='same')(up6)
conv6 = Conv2D(256, (3, 3), activation='relu', padding='same')(conv6)
up_conv6 = Conv2DTranspose(128, (2, 2), strides=(2, 2), padding='same')(conv6)
ch, cw = get_crop_shape(conv3, up_conv6)
crop_conv3 = Cropping2D(cropping=(ch, cw), data_format="channels_last")(conv3)
up7 = concatenate([up_conv6, crop_conv3], axis=3)
conv7 = Conv2D(128, (3, 3), activation='relu', padding='same')(up7)
conv7 = Conv2D(128, (3, 3), activation='relu', padding='same')(conv7)
up_conv7 = Conv2DTranspose(64, (2, 2), strides=(2, 2), padding='same')(conv7)
ch, cw = get_crop_shape(conv2, up_conv7)
crop_conv2 = Cropping2D(cropping=(ch, cw), data_format="channels_last")(conv2)
up8 = concatenate([up_conv7, crop_conv2], axis=3)
conv8 = Conv2D(64, (3, 3), activation='relu', padding='same')(up8)
conv8 = Conv2D(64, (3, 3), activation='relu', padding='same')(conv8)
up_conv8 = Conv2DTranspose(64, (2, 2), strides=(2, 2), padding='same')(conv8)
ch, cw = get_crop_shape(conv1, up_conv8)
crop_conv1 = Cropping2D(cropping=(ch, cw), data_format="channels_last")(conv1)
up9 = concatenate([up_conv8, crop_conv1], axis=3)
conv9 = Conv2D(32, (3, 3), activation='relu', padding='same')(up9)
conv9 = Conv2D(32, (3, 3), activation='relu', padding='same')(conv9)
conv10 = Conv2D(1, (1, 1), activation='sigmoid')(conv9)
model = Model(inputs=[inputs], outputs=[conv10])
model.compile(optimizer=Adam(lr=1e-5), loss=dice_coef_loss, metrics=[dice_coef])
return model
IMG_WIDTH = 512
IMG_HEIGHT = 512
IMG_CHANNELS = 1
outputPath = 'train_liver.h5'
val_outputPath = 'val_liver.h5'
BATCH_SIZE = 8
BATCH_SIZE就是每个BATCH训练多少张图片,根据电脑性能调整,设的太大可能跑不出来,而且还会影响steps_per_epoch,会对训练结果产生影响,所以谨慎修改。
class UnetModel:
def train_and_predict(self):
reader = HDF5DatasetGenerator(dbPath=outputPath,batchSize=BATCH_SIZE)
train_iter = reader.generator()
test_reader = HDF5DatasetGenerator(dbPath=val_outputPath,batchSize=BATCH_SIZE)
test_iter = test_reader.generator()
for zf in range(int(VAL_TOTAL/BATCH_SIZE)+1):#如果不加这个循环,每次只能跑出BATCH_SIZE张图片
fixed_test_images, fixed_test_masks = test_iter.__next__()
model = get_unet()
model_checkpoint = ModelCheckpoint('weights.h5', monitor='val_loss', save_best_only=True)
model.fit_generator(train_iter, steps_per_epoch=TOTAL/BATCH_SIZE, verbose=1, epochs=20, shuffle=True,
validation_data=(fixed_test_images, fixed_test_masks), callbacks=[model_checkpoint])
print('-' * 30)
print('Loading and preprocessing test data...')
print('-' * 30)
print('-' * 30)
print('Loading saved weights...')
print('-' * 30)
model.load_weights('weights.h5')
print('-' * 30)
print('Predicting masks on test data...')
print('-' * 30)
#print(fixed_test_images.shape)
imgs_mask_test = model.predict(fixed_test_images, verbose=1)
np.save('imgs_mask_test.npy', imgs_mask_test)
print('-' * 30)
print('Saving predicted masks to files...')
print('-' * 30)
pred_dir = 'preds'
if not os.path.exists(pred_dir):
os.mkdir(pred_dir)
i = 0
print(imgs_mask_test.shape)
for image in imgs_mask_test:
image = (image[:, :, 0] * 255.).astype(np.uint8)
gt = (fixed_test_masks[i, :, :, 0] * 255.).astype(np.uint8)
ini = (fixed_test_images[i, :, :, 0] * 255.).astype(np.uint8)
io.imsave(os.path.join(pred_dir, str(i+ zf*BATCH_SIZE) + '_ini.png'), ini)
io.imsave(os.path.join(pred_dir, str(i+ zf*BATCH_SIZE) + '_pred.png'), image)
io.imsave(os.path.join(pred_dir, str(i+ zf*BATCH_SIZE) + '_gt.png'), gt)
i += 1
reader.close()
test_reader.close()
unet = UnetModel()
unet.train_and_predict()
读dcm的问题
在这个项目中读dcm总是出现各种各样的问题,可能是这个项目给我们提供的dcm图片格式有点问题,导致用pydicom读的时候有些属性没有,而用其他dcm读却正常。所以最后我们采用了SimpleITK来读取,但是读出来和pydicom读出来的属性是不一样的,这一点需要注意,可以将两种方式读取出来的属性一一对应,将用pydicom读出来的变量转换成用SimpleITK读出来的变量,程序就能正常运行了。
数据说明
 里面有两个文件夹arterial phase放CT图,mask放掩模图
里面有两个文件夹arterial phase放CT图,mask放掩模图
 这是掩模图的文件夹,都是png文件,注意相对应的CT图和掩模图再文件夹里的位置一定要对应。
这是掩模图的文件夹,都是png文件,注意相对应的CT图和掩模图再文件夹里的位置一定要对应。
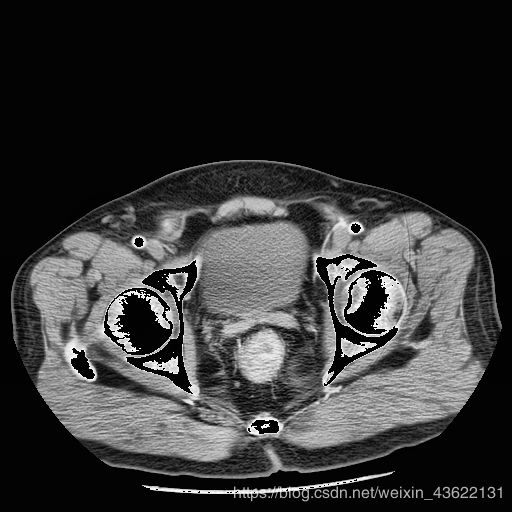
结果展示
个别部分的棱角可能不太明显,但是整体形状以及位置是很相近的,而且我们只用了18个人做训练集,这个预测的模型用的是准确率到0.8的模型,经过参数的调整还能达到更高,大家可以再进行调试,一定可以得出更好的结果。
鸣谢
这个代码的整体框架是借鉴医学图像分割 基于深度学习的肝脏肿瘤分割 实战(一)这篇博客的,这篇关于Unet分割肝脏肿瘤的文章也是很棒的,大家也可以进行参考。