The impact of different initialization strategies on neural
This article first appeared in ai-exception
Abstract
As we know, a well chosen initialization can speed up the convergence of gradient descent and increase the odds of gradient descent converging to a lower training (and generalization) error .So in this article, we will explore the impact of different parameter initialization strategies on training in deep learning.
In this experience,we will try three different initialization strategies:
- Zeros initialization
- Random initialization
- He initialization
Load dataset
To get started, we need load dataset for our experience :
import numpy as np
import matplotlib.pyplot as plt
import sklearn
import sklearn.datasets
from init_utils import sigmoid, relu, compute_loss, forward_propagation, backward_propagation
from init_utils import update_parameters, predict, load_dataset, plot_decision_boundary, predict_dec
plt.rcParams['figure.figsize'] = (7.0, 4.0) # set default size of plots
plt.rcParams['image.interpolation'] = 'nearest'
plt.rcParams['image.cmap'] = 'gray'
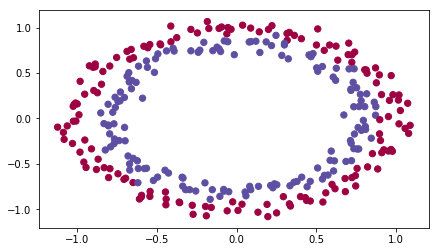
# load image dataset: blue/red dots in circles
train_X, train_Y, test_X, test_Y = load_dataset()
plt.show()
What we goal is train a model toi classifier the blue dots and red dots.
Neural Network model
I will use a 3-layer neural network (already implemented on init_utils). Here are the initialization methods we will experiment with:
- Zeros initialization – setting
initialization = "zeros"in the input argument. - Random initialization – setting
initialization = "random"in the input argument. This initializes the weights to large random values. - He initialization – setting
initialization = "he"in the input argument. This initializes the weights to random values scaled according to a paper by He et al., 2015.
The model code as follows:
def model(X, Y, learning_rate = 0.01, num_iterations = 15000, print_cost = True, initialization = "he"):
"""
Implements a three-layer neural network: LINEAR->RELU->LINEAR->RELU->LINEAR->SIGMOID.
Arguments:
X -- input data, of shape (2, number of examples)
Y -- true "label" vector (containing 0 for red dots; 1 for blue dots), of shape (1, number of examples)
learning_rate -- learning rate for gradient descent
num_iterations -- number of iterations to run gradient descent
print_cost -- if True, print the cost every 1000 iterations
initialization -- flag to choose which initialization to use ("zeros","random" or "he")
Returns:
parameters -- parameters learnt by the model
"""
grads = {}
costs = [] # to keep track of the loss
m = X.shape[1] # number of examples
layers_dims = [X.shape[0], 10, 5, 1]
# Initialize parameters dictionary.
if initialization == "zeros":
parameters = initialize_parameters_zeros(layers_dims)
elif initialization == "random":
parameters = initialize_parameters_random(layers_dims)
elif initialization == "he":
parameters = initialize_parameters_he(layers_dims)
# Loop (gradient descent)
for i in range(0, num_iterations):
# Forward propagation: LINEAR -> RELU -> LINEAR -> RELU -> LINEAR -> SIGMOID.
a3, cache = forward_propagation(X, parameters)
# Loss
cost = compute_loss(a3, Y)
# Backward propagation.
grads = backward_propagation(X, Y, cache)
# Update parameters.
parameters = update_parameters(parameters, grads, learning_rate)
# Print the loss every 1000 iterations
if print_cost and i % 1000 == 0:
print("Cost after iteration {}: {}".format(i, cost))
costs.append(cost)
# plot the loss
plt.figure()
plt.plot(costs)
plt.ylabel('cost')
plt.xlabel('iterations (per hundreds)')
plt.title('init with' initialization ' Learning rate =' str(learning_rate))
plt.show()
return parameters
2 - Zero initialization
There are two types of parameters to initialize in a neural network:
- the weight matrices ( W [ 1 ] , W [ 2 ] , W [ 3 ] , . . . , W [ L − 1 ] , W [ L ] ) (W^{[1]}, W^{[2]}, W^{[3]}, ..., W^{[L-1]}, W^{[L]}) (W[1],W[2],W[3],...,W[L−1],W[L])
- the bias vectors ( b [ 1 ] , b [ 2 ] , b [ 3 ] , . . . , b [ L − 1 ] , b [ L ] ) (b^{[1]}, b^{[2]}, b^{[3]}, ..., b^{[L-1]}, b^{[L]}) (b[1],b[2],b[3],...,b[L−1],b[L])
Exercise: Implement the following function to initialize all parameters to zeros. You’ll see later that this does not work well since it fails to “break symmetry”, but lets try it anyway and see what happens. Use np.zeros((…,…)) with the correct shapes.
def initialize_parameters_zeros(layers_dims):
"""
Arguments:
layer_dims -- python array (list) containing the size of each layer.
Returns:
parameters -- python dictionary containing your parameters "W1", "b1", ..., "WL", "bL":
W1 -- weight matrix of shape (layers_dims[1], layers_dims[0])
b1 -- bias vector of shape (layers_dims[1], 1)
...
WL -- weight matrix of shape (layers_dims[L], layers_dims[L-1])
bL -- bias vector of shape (layers_dims[L], 1)
"""
parameters = {}
L = len(layers_dims) # number of layers in the network
for l in range(1, L):
parameters['W' str(l)] = np.zeros(shape=(layers_dims[l], layers_dims[l - 1]))
parameters['b' str(l)] = np.zeros(shape=(layers_dims[l], 1))
return parameters
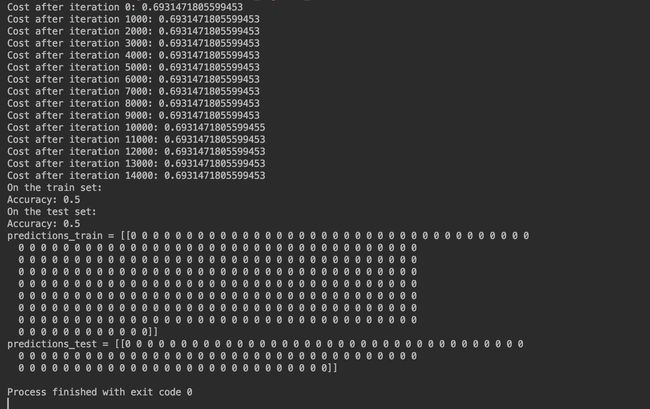
Run the following code to train our model on 15,000 iterations using zeros initialization:
def train_with_zeros_init():
parameters = model(train_X, train_Y, initialization="zeros")
print("On the train set:")
predictions_train = predict(train_X, train_Y, parameters)
print("On the test set:")
predictions_test = predict(test_X, test_Y, parameters)
print("predictions_train = " str(predictions_train))
print("predictions_test = " str(predictions_test))
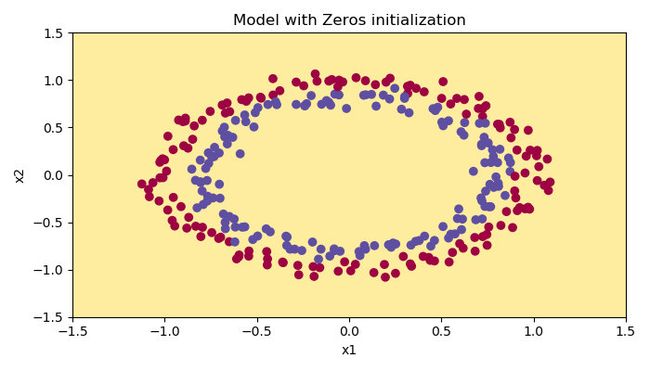
plt.figure()
plt.title("Model with Zeros initialization")
axes = plt.gca()
axes.set_xlim([-1.5, 1.5])
axes.set_ylim([-1.5, 1.5])
plot_decision_boundary(lambda x: predict_dec(parameters, x.T), train_X, train_Y)
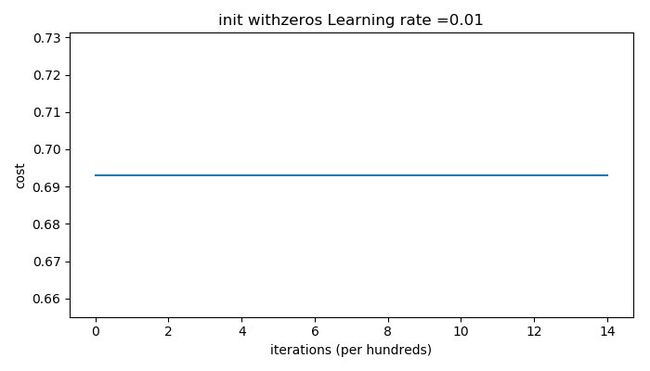
The outputs are as follows:
The performance is really bad, and the cost does not really decrease, and the algorithm performs no better than random guessing. Why? From the details of the predictions and the decision boundary, we can find that the model is predicting 0 for every example.
In general, initializing all the weights to zero results in the network failing to break symmetry(对称性). This means that every neuron in each layer will learn the same thing, and you might as well be training a neural network with n [ l ] = 1 n^{[l]}=1 n[l]=1 for every layer, and the network is no more powerful than a linear classifier such as logistic regression.
What we should remember
- The weights W [ l ] W^{[l]} W[l] should be initialized randomly to break symmetry.
- It is however okay to initialize the biases b [ l ] b^{[l]} b[l] to zeros. Symmetry is still broken so long as W [ l ] W^{[l]} W[l] is initialized randomly.
Random initialization
To break symmetry, lets intialize the weights randomly. Following random initialization, each neuron can then proceed to learn a different function of its inputs. In this exercise, you will see what happens if the weights are intialized randomly, but to very large values.
def initialize_parameters_random(layers_dims):
"""
Arguments:
layer_dims -- python array (list) containing the size of each layer.
Returns:
parameters -- python dictionary containing your parameters "W1", "b1", ..., "WL", "bL":
W1 -- weight matrix of shape (layers_dims[1], layers_dims[0])
b1 -- bias vector of shape (layers_dims[1], 1)
...
WL -- weight matrix of shape (layers_dims[L], layers_dims[L-1])
bL -- bias vector of shape (layers_dims[L], 1)
"""
np.random.seed(3) # This seed makes sure your "random" numbers will be the as ours
parameters = {}
L = len(layers_dims) # integer representing the number of layers
for l in range(1, L):
parameters['W' str(l)] = np.random.randn(layers_dims[l], layers_dims[l - 1])*10
parameters['b' str(l)] = np.zeros(shape=(layers_dims[l], 1))
return parameters
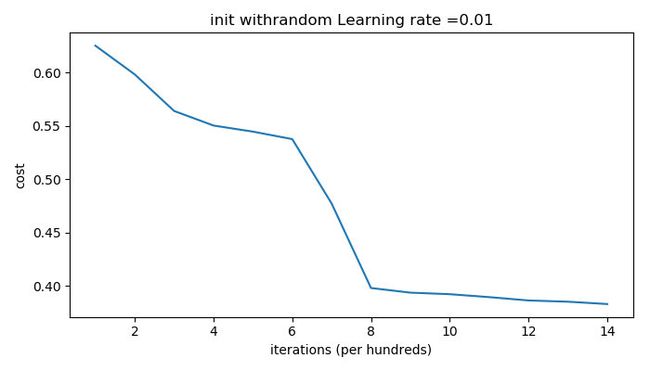
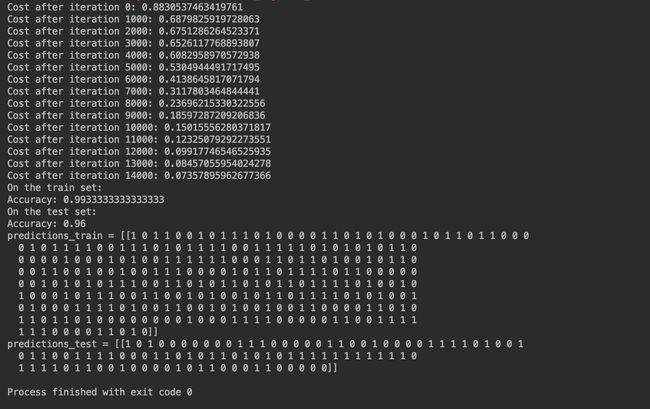
Run the following code to train our model on 15,000 iterations using random initialization:
def train_with_random_init():
parameters = model(train_X, train_Y, initialization="random")
print("On the train set:")
predictions_train = predict(train_X, train_Y, parameters)
print("On the test set:")
predictions_test = predict(test_X, test_Y, parameters)
print("predictions_train = " str(predictions_train))
print("predictions_test = " str(predictions_test))
plt.figure()
plt.title("Model with random initialization")
axes = plt.gca()
axes.set_xlim([-1.5, 1.5])
axes.set_ylim([-1.5, 1.5])
plot_decision_boundary(lambda x: predict_dec(parameters, x.T), train_X, train_Y)
And the outputs are as follows:
We can see “inf” as the cost after the iteration 0, this is because of numerical roundoff; a more numerically sophisticated implementation would fix this. But this isn’t worth worrying about for our purposes.
Anyway, it looks like we have broken symmetry, and this gives better results. than before. The model is no longer outputting all 0s.
Observations:
- The cost starts very high. This is because with large random-valued weights, the last activation (sigmoid) outputs results that are very close to 0 or 1 for some examples, and when it gets that example wrong it incurs a very high loss for that example. Indeed, when log ( a [ 3 ] ) = log ( 0 ) \log(a^{[3]}) = \log(0) log(a[3])=log(0), the loss goes to infinity.
- Poor initialization can lead to vanishing/exploding gradients, which also slows down the optimization algorithm.
- If you train this network longer you will see better results, but initializing with overly large random numbers slows down the optimization.
In summary
- Initializing weights to very large random values does not work well.
- Hopefully intializing with small random values does better. The important question is: how small should be these random values be? Lets find out in the next part!
He initialization
Finally, try “He Initialization”; this is named for the first author of He et al., 2015. (If you have heard of “Xavier initialization”, this is similar except Xavier initialization uses a scaling factor for the weights W [ l ] W^{[l]} W[l] of sqrt(1./layers_dims[l-1]) where He initialization would use sqrt(2./layers_dims[l-1]).):
def initialize_parameters_he(layers_dims):
"""
Arguments:
layer_dims -- python array (list) containing the size of each layer.
Returns:
parameters -- python dictionary containing your parameters "W1", "b1", ..., "WL", "bL":
W1 -- weight matrix of shape (layers_dims[1], layers_dims[0])
b1 -- bias vector of shape (layers_dims[1], 1)
...
WL -- weight matrix of shape (layers_dims[L], layers_dims[L-1])
bL -- bias vector of shape (layers_dims[L], 1)
"""
np.random.seed(3)
parameters = {}
L = len(layers_dims) - 1 # integer representing the number of layers
for l in range(1, L 1):
parameters['W' str(l)] = np.random.randn(layers_dims[l], layers_dims[l - 1]) * np.sqrt(2 / layers_dims[l - 1])
parameters['b' str(l)] = np.zeros(shape=(layers_dims[l], 1))
return parameters
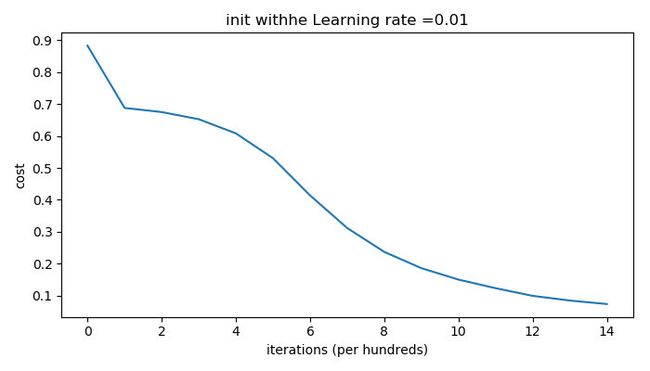
And run the code as follow to train with he initialization:
def train_with_he_init():
parameters = model(train_X, train_Y, initialization="he")
print("On the train set:")
predictions_train = predict(train_X, train_Y, parameters)
print("On the test set:")
predictions_test = predict(test_X, test_Y, parameters)
print("predictions_train = " str(predictions_train))
print("predictions_test = " str(predictions_test))
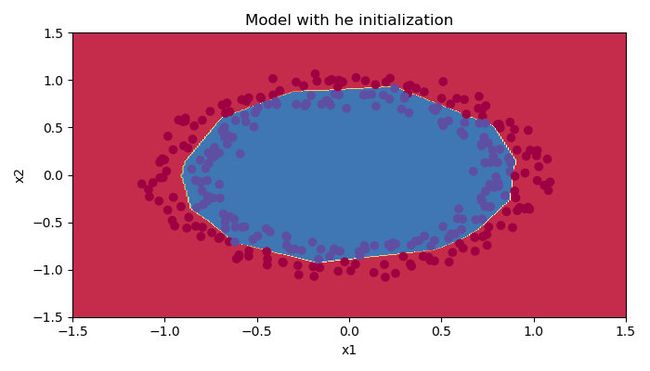
plt.figure()
plt.title("Model with he initialization")
axes = plt.gca()
axes.set_xlim([-1.5, 1.5])
axes.set_ylim([-1.5, 1.5])
plot_decision_boundary(lambda x: predict_dec(parameters, x.T), train_X, train_Y)
The outputs are as follows:
Observations:
- The model with He initialization separates the blue and the red dots very well in a small number of iterations.
Conclusions
We have seen three different types of initializations. For the same number of iterations and same hyperparameters the comparison is:
| Model | Train accuracy | Problem/Comment |
| 3-layer NN with zeros initialization | 50% | fails to break symmetry |
| 3-layer NN with large random initialization | 83% | too large weights |
| 3-layer NN with He initialization | 99% | recommended method |
What we should remember from this blog
- Different initializations lead to different results
- Random initialization is used to break symmetry and make sure different hidden units can learn different things
- Don’t intialize to values that are too large
- He initialization works well for networks with ReLU activations.
links
[github