机器学习随笔6--支持向量机
摘要
本文是作者最近在学习支持向量机的随笔,主要是简单介绍支持向量机,阐述支持向量机的原理、SMO算法、核函数以及实际数据测试效果,附录含有代码。
目录
- 摘要
- 目录
- 一、支持向量机简介
- 二、支持向量机的原理
- 2.1 二维的样本空间
- 2.2 高维的样本空间
- 2.3 对偶问题
- 三、SMO算法
- 四、核函数
- 五、测试
- 5.1 线性可分数据
- 5.2 非线性可分数据
- 六、小结
- 七、参考文献
- 八、附录
一、支持向量机简介
支持向量机(Support Vector Machines,SVM)是一个二类分类模型,它的基本模型是定义在特征空间上的间隔最大的线性分类器。直观理解是,支持向量机的目标是寻找一个超平面将待测数据分成两种类别。
二、支持向量机的原理
2.1 二维的样本空间
现在考虑二维情况,有一个样本数为n,标签为二类的二维数据集:
{(x(1)1,x(2)1;y1),(x(1)2,x(2)2;y2),…,(x(1)n,x(2)n;yn)}
其中,x(1)n,x(2)n分别指第i个数据的第一个、第二个分量,yi指第i个数据的标签,并且yi∈{1,−1}。
可视化即是:
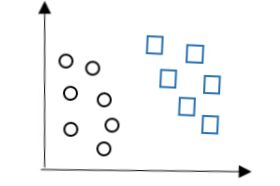
图1
现在的任务是在二维平面上画出一条直线将上图两类数据分隔开。这分法可就多了,我们可以画出的直线可以有红色、绿色、灰色三条直线。
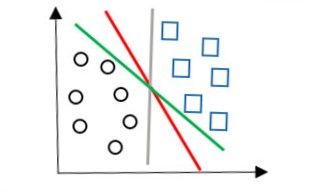
图2
到底哪种分法最好呢?明显看出,红色的直线划分的最好。因为红色直线跟两种类别的每一个数据点的距离都很适合,跟圆圈的最短距离比绿色直线的大,跟方形的最短距离比灰色直线的大。也就是说,这样的红色直线到不同类别的距离都是最大的。
上述例子是在二维平面中寻找超平面,直接用肉眼就可以抉择,然而如果是三维或者更高维的情况,单凭人类肉眼是无能为力的,但我们是可以通过计算机来寻找啊,通过相应的数学知识,建立对应的数学模型,通过计算机来求解。
2.2 高维的样本空间
给定n个训练样本:{(x1,y1),(x2,y2),...,(xn,yn)} ,其中xi是d维向量,表明了每个样本具有d个属性;yi指的是类别,并且 yi∈{1,−1}。
(1)在样本空间中,划分超平面P={x|f(x)=α,x∈Rd,α∈R}可通过线性方程wTx+b=0 来描述,要计算样本点A到划分超平面P的距离,要给出点到划分超平面P的法线或者垂线的长度,该值为:
d(A,P)=|wTA+b|||w||
事实上,可知所有样本点属于Rd,因此是希尔伯特空间,则对于点A,存在点B∈P,使得||AB||=d(A,P),而且AB⊥P。
又知w||w||是超平面P的一个单位法向量,则有
AB||AB||=±w||w||
则
⟨w||w||,AB||AB||⟩=±⟨w||w||,w||w||⟩
由于
⟨w||w||,w||w||⟩=1
则
||AB||=\pm \frac{\left< w,AB \right>}{||w||}=\pm \frac{\left< w,A-B \right>}{||w||}
若AB与w同向,
||AB||= \frac{\left< w,A-B \right>}{||w||}= \frac{w^TA-w^TB}{||w||} = \frac{w^TA+b}{||w||}
否则,
||AB||=- \frac{\left< w,A-B \right>}{||w||}=- \frac{w^TA-w^TB}{||w||} =-\frac{w^TA+b}{||w||}
综上,
d(A,P)=||AB||=\frac{|w^TA+b|}{||w||}
于是,命题得证。
可知,向量w 和常数b 一起描述所给数据的超平面。
(2)假设超平面能够正确分类样本,则可以通过对w缩放可以使得下式成立
y_i=1 :w^Tx_i+b \geqslant 1
y_i=-1 :w^Tx_i+b \leqslant -1
距离超平面最近的几个样本点使得上式等号成立,称作“支持向量”。两个异类支持向量到超平面的距离之和称为“间隔”。
借图来表示就是:
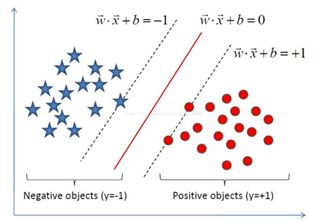
图3
首先,间隔的公式为:
d(x_i^+,P)+d(x_i^-,P)= \frac {|1|}{||w||}+ \frac {|-1|}{||w||}= \frac {2}{||w||}
目标是使得间隔最大,即是:
\max_{w,b} \frac{2}{||w||} \Longleftrightarrow \min_{w,b} \frac{1}{2}||w||^2
在超平面两侧,则有:
y_i(w^Tx_i+b) \geqslant 1,i=1,2,...,n
(3)支持向量机的数学模型为:
\min_{w,b} \frac{1}{2}||w||^2\\ s.t. \ y_i(w^Tx_i+b) \geqslant 1,i=1,2,...,n
2.3 对偶问题
上述问题是一个凸二次规划问题,在满足KKT(Karush-Kuhn-Tucker)条件,可以通过拉格朗日乘子法变换到对偶变量的优化问题之后,可以找到一种更加有效的方法来进行求解。
(1)原问题的拉格朗日函数为(其中,\alpha_i \geqslant 0是拉格朗日乘子):
L(w,b,\alpha)= \frac{1}{2} ||w||^2 + \sum_{i=1}^n \alpha_i(1-y_i(w^Tx_i+b))
(2)需要满足的KKT条件是:
\left\{ \begin{array}{lr} \alpha_i \geqslant 0 \\ y_i(w^Tx_i+b )-1 \geqslant 0 \\ \alpha_i \cdot ( y_i (w^Tx_i+b )-1 )= 0 \end{array} \right.
(3)为求得其对偶问题
令其偏导数等于0,即
\frac { \partial L}{\partial w}=\frac { \partial L}{\partial b}=0
这里用到向量和矩阵求导的知识,对于向量函数x、常向量v、矩阵A:
\frac { \partial (v^T x) }{\partial x}=v \\ \frac { \partial (x^T A x) }{\partial x}=(A+A^T)x
对w有
||w||^2= \left< w,w \right>=w^T E w
所以
\frac{1}{2} \frac { \partial }{\partial w} ||w||^2 = \frac{1}{2} \partial (w^T Ew)= \frac{1}{2} (E+E^T)w=w
同样可得
\frac { \partial }{\partial w} \sum_{i=1}^n \alpha_i(1-y_i(w^Tx_i+b))= -\sum _{i=1}^n \alpha_i y_i x_i
则得:
\frac { \partial L}{\partial w}=w-\sum _{i=1}^n \alpha_i y_i x_i =0
所以对w有:
w=\sum _{i=1}^n \alpha_i y_i x_i
另一方面,令b的偏导数等于0,即是:
\sum_{i=1} ^ n \alpha _i y_i =0
代入拉格朗日函数得对偶问题:
\max_{\alpha} \sum_{i=1}^n \alpha_i - \frac{1}{2} \sum _{i=1}^n \sum_{j=1}^n \alpha_i \alpha_j y_i y_j x_i^Tx_j
s.t. \left\{ \begin{gather*} \sum_{i=1} ^ {n} \alpha _i y_i = 0 \\ \alpha _i \geqslant 0 ,i =1,2,...,n \end{gather*} \right.
解出所有的\alpha_i之后,首先求出w,即是:
w=\sum _{i=1}^n \alpha_i y_i x_i
利用w^Tx_i+b= \pm 1的数据点,即“支持向量”,求出b得:
b= -\frac{\max \{w^Tx_i|y_i=-1\}+\min\{w^Tx_i|y_i=1\} }{2}
即可得到划分模型为:
\begin{align*} f(x)& =w^Tx+b\\ & =\sum_{i=1}^{n} \alpha_iy_ix_i^Tx+b \end{align*}
三、SMO算法
步骤:
不断执行以下两个步骤直到收敛
(1)选取一对需要更新的变量\alpha_i和\alpha_j;
(2)固定\alpha_i和\alpha_j以外的参数,求解对偶问题更新后的\alpha_i和\alpha_j。
只要选取的\alpha_i和\alpha_j中有一个不满足KKT条件, 目标函数就会在迭代后减小。KKT条件违背的程度越大,变量更新后可能导致的目标函数值减幅越大。
使选取的两变量所对应样本之间的间隔最大(两个变量有很大的差别,对它们进行更新会带给目标函数值更大的变化)。
四、核函数
原始样本空间线性不可分时,样本从原始空间作一映射\varPhi到一个更高维的特征空间,使得样本在这个特征空间内线性可分。如果原始空间是有限维,那么一定存在一个高维特征空间使样本可分。
高维的特征空间中,划分超平面变成:
w^T \varPhi (x) +b=0
此时,支持向量机的模型为:
\min_{w,b} \frac{1}{2}||w||^2\\ s.t. \ y_i(w^T \varPhi (x_i)+b) \geqslant 1,i=1,2,...,n
同理可得,其对偶问题为:
\max_{\alpha} \sum_{i=1}^n \alpha_i -\frac{1}{2} \sum _{i=1}^n \sum_{j=1}^n \alpha_i \alpha_j y_i y_j \varPhi (x_i)^T \varPhi (x_j)
s.t. \left\{ \begin{gather*} \sum_{i=1} ^ {n} \alpha _i y_i = 0 \\ \alpha _i \geqslant 0 ,i =1,2,...,n \end{gather*} \right.
由于特征空间维数可能很高,直接计算\varPhi (x_i)^T \varPhi (x_j) 通常是困难的。设想函数k(x_i,x_j)=\varPhi (x_i)^T \varPhi (x_j) , 在特征空间x_i,x_j的内积等于它们在原始样本空间中通过核函数计算的结果。
在此引入径向基核函数,它是一个采用向量作为自变量的函数,能够基于向量距离运算输出一个标量。常用核函数有:
(1)线性核
k(x_i,x_j)=x_i^Tx_j
可知上述线性核就是在线性可分数据中支持向量机使用的内积。
(2)多项式核
k(x_i,x_j)=( x_i^Tx_j ) ^m
(3)高斯核
k(x_i,x_j)=\exp ( \frac { -||x_i-x_j||^2}{2\sigma ^2})
(4)拉普拉斯核
k(x_i,x_j)=\exp ( \frac { -||x_i-x_j||}{\sigma })
五、测试
5.1 线性可分数据
在此使用一个标签为二元(即1与-1)的属性为2个的数据集进行支持向量机的测试,以验证支持向量机的运行情况。其中,原始数据如图所示,明显看出次数据集是线性可分的,而分割线应该是显然可知的。
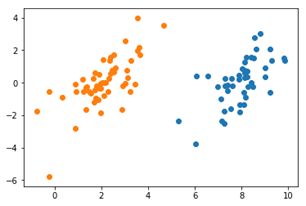
图4
使用SMO算法求解得出:
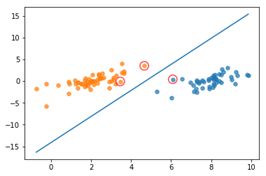
图5
5.2 非线性可分数据
使用非线性可分的数据集,如图所示。但明显可以看出该数据集有一个可以分类的模式,即可以使用某个圆把一个数据囊括。
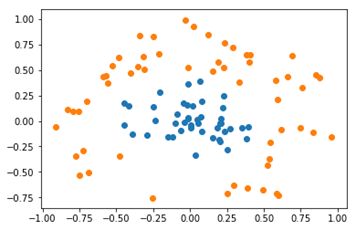
图6
使用径向基核函数的高斯版本进行计算,结果为:

图7
可知,其支持向量个数为28,测试仅有5%错误率。
六、小结
支持向量机是一种通过求解凸二次规划问题来解决分类问题的算法,具有较低的泛化错误率。而SMO算法可以通过每次只优化\alpha_i和\alpha_j值来加快SVM的训练速度。
核技巧是将数据由低维空间映射到高维空间,可以将一个低维空间中的非线性问题转换为高维空间下的线性问题来求解。而径向基核函数是一个常用的度量两个向量距离的核函数。
最后,支持向量机的优缺点:
(1)优点:泛化错误率低,计算开销不大
(2)缺点:对参数调节和核函数的选择敏感,且仅适用于二类分类
七、参考文献
[1]周志华.机器学习[M].北京:清华大学出版社,2016.
[2]Peter Harrington.机器学习实战[M].北京:人民邮电出版社,2013.
[3]韩家炜等.数据挖掘概念与技术[M].北京:机械工业出版社,2012.
八、附录
在此借用《机器学习实战》中的代码:
其中,svmMLiA.py文件为:
"""
Created on Sat May 5 23:56:01 2018
@author: Diky
"""
"""
_oo0oo_
o8888888o
88" . "88
(| -_- |)
0\ = /0
___/`---'\___
.' \\| |// '.
/ \\||| : |||// \
/ _||||| -:- |||||- \
| | \\\ - /// | |
| \_| ''\---/'' |_/ |
\ .-\__ '-' ___/-. /
___'. .' /--.--\ `. .'___
."" '< `.___\_<|>_/___.' >' "".
| | : `- \`.;`\ _ /`;.`/ - ` : | |
\ \ `_. \_ __\ /__ _/ .-` / /
=====`-.____`.___ \_____/___.-`___.-'=====
`=---='
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
佛祖保佑 永无BUG
"""
from numpy import *
from time import sleep
def loadDataSet(fileName):
dataMat = []; labelMat = []
fr = open(fileName)
for line in fr.readlines():
lineArr = line.strip().split('\t')
dataMat.append([float(lineArr[0]), float(lineArr[1])])
labelMat.append(float(lineArr[2]))
return dataMat,labelMat
def selectJrand(i,m):
j=i #we want to select any J not equal to i
while (j==i):
j = int(random.uniform(0,m))
return j
def clipAlpha(aj,H,L):
if aj > H:
aj = H
if L > aj:
aj = L
return aj
def smoSimple(dataMatIn, classLabels, C, toler, maxIter):
dataMatrix = mat(dataMatIn); labelMat = mat(classLabels).transpose()
b = 0; m,n = shape(dataMatrix)
alphas = mat(zeros((m,1)))
iter = 0
while (iter < maxIter):
alphaPairsChanged = 0
for i in range(m):
fXi = float(multiply(alphas,labelMat).T*(dataMatrix*dataMatrix[i,:].T)) + b
Ei = fXi - float(labelMat[i])#if checks if an example violates KKT conditions
if ((labelMat[i]*Ei < -toler) and (alphas[i] < C)) or ((labelMat[i]*Ei > toler) and (alphas[i] > 0)):
j = selectJrand(i,m)
fXj = float(multiply(alphas,labelMat).T*(dataMatrix*dataMatrix[j,:].T)) + b
Ej = fXj - float(labelMat[j])
alphaIold = alphas[i].copy(); alphaJold = alphas[j].copy();
if (labelMat[i] != labelMat[j]):
L = max(0, alphas[j] - alphas[i])
H = min(C, C + alphas[j] - alphas[i])
else:
L = max(0, alphas[j] + alphas[i] - C)
H = min(C, alphas[j] + alphas[i])
if L==H: print ("L==H"); continue
eta = 2.0 * dataMatrix[i,:]*dataMatrix[j,:].T - dataMatrix[i,:]*dataMatrix[i,:].T - dataMatrix[j,:]*dataMatrix[j,:].T
if eta >= 0: print ("eta>=0"); continue
alphas[j] -= labelMat[j]*(Ei - Ej)/eta
alphas[j] = clipAlpha(alphas[j],H,L)
if (abs(alphas[j] - alphaJold) < 0.00001): print ("j not moving enough"); continue
alphas[i] += labelMat[j]*labelMat[i]*(alphaJold - alphas[j])#update i by the same amount as j
#the update is in the oppostie direction
b1 = b - Ei- labelMat[i]*(alphas[i]-alphaIold)*dataMatrix[i,:]*dataMatrix[i,:].T - labelMat[j]*(alphas[j]-alphaJold)*dataMatrix[i,:]*dataMatrix[j,:].T
b2 = b - Ej- labelMat[i]*(alphas[i]-alphaIold)*dataMatrix[i,:]*dataMatrix[j,:].T - labelMat[j]*(alphas[j]-alphaJold)*dataMatrix[j,:]*dataMatrix[j,:].T
if (0 < alphas[i]) and (C > alphas[i]): b = b1
elif (0 < alphas[j]) and (C > alphas[j]): b = b2
else: b = (b1 + b2)/2.0
alphaPairsChanged += 1
print ("iter: %d i:%d, pairs changed %d" % (iter,i,alphaPairsChanged))
if (alphaPairsChanged == 0): iter += 1
else: iter = 0
print ("iteration number: %d" % iter)
return b,alphas
def kernelTrans(X, A, kTup): #calc the kernel or transform data to a higher dimensional space
m,n = shape(X)
K = mat(zeros((m,1)))
if kTup[0]=='lin': K = X * A.T #linear kernel
elif kTup[0]=='rbf':
for j in range(m):
deltaRow = X[j,:] - A
K[j] = deltaRow*deltaRow.T
K = exp(K/(-1*kTup[1]**2)) #divide in NumPy is element-wise not matrix like Matlab
else: raise NameError('Houston We Have a Problem -- \
That Kernel is not recognized')
return K
class optStruct:
def __init__(self,dataMatIn, classLabels, C, toler, kTup): # Initialize the structure with the parameters
self.X = dataMatIn
self.labelMat = classLabels
self.C = C
self.tol = toler
self.m = shape(dataMatIn)[0]
self.alphas = mat(zeros((self.m,1)))
self.b = 0
self.eCache = mat(zeros((self.m,2))) #first column is valid flag
self.K = mat(zeros((self.m,self.m)))
for i in range(self.m):
self.K[:,i] = kernelTrans(self.X, self.X[i,:], kTup)
def calcEk(oS, k):
fXk = float(multiply(oS.alphas,oS.labelMat).T*oS.K[:,k] + oS.b)
Ek = fXk - float(oS.labelMat[k])
return Ek
def selectJ(i, oS, Ei): #this is the second choice -heurstic, and calcs Ej
maxK = -1; maxDeltaE = 0; Ej = 0
oS.eCache[i] = [1,Ei] #set valid #choose the alpha that gives the maximum delta E
validEcacheList = nonzero(oS.eCache[:,0].A)[0]
if (len(validEcacheList)) > 1:
for k in validEcacheList: #loop through valid Ecache values and find the one that maximizes delta E
if k == i: continue #don't calc for i, waste of time
Ek = calcEk(oS, k)
deltaE = abs(Ei - Ek)
if (deltaE > maxDeltaE):
maxK = k; maxDeltaE = deltaE; Ej = Ek
return maxK, Ej
else: #in this case (first time around) we don't have any valid eCache values
j = selectJrand(i, oS.m)
Ej = calcEk(oS, j)
return j, Ej
def updateEk(oS, k):#after any alpha has changed update the new value in the cache
Ek = calcEk(oS, k)
oS.eCache[k] = [1,Ek]
def innerL(i, oS):
Ei = calcEk(oS, i)
if ((oS.labelMat[i]*Ei < -oS.tol) and (oS.alphas[i] < oS.C)) or ((oS.labelMat[i]*Ei > oS.tol) and (oS.alphas[i] > 0)):
j,Ej = selectJ(i, oS, Ei) #this has been changed from selectJrand
alphaIold = oS.alphas[i].copy(); alphaJold = oS.alphas[j].copy();
if (oS.labelMat[i] != oS.labelMat[j]):
L = max(0, oS.alphas[j] - oS.alphas[i])
H = min(oS.C, oS.C + oS.alphas[j] - oS.alphas[i])
else:
L = max(0, oS.alphas[j] + oS.alphas[i] - oS.C)
H = min(oS.C, oS.alphas[j] + oS.alphas[i])
if L==H: print( "L==H"); return 0
eta = 2.0 * oS.K[i,j] - oS.K[i,i] - oS.K[j,j] #changed for kernel
if eta >= 0: print ("eta>=0"); return 0
oS.alphas[j] -= oS.labelMat[j]*(Ei - Ej)/eta
oS.alphas[j] = clipAlpha(oS.alphas[j],H,L)
updateEk(oS, j) #added this for the Ecache
if (abs(oS.alphas[j] - alphaJold) < 0.00001): print( "j not moving enough"); return 0
oS.alphas[i] += oS.labelMat[j]*oS.labelMat[i]*(alphaJold - oS.alphas[j])#update i by the same amount as j
updateEk(oS, i) #added this for the Ecache #the update is in the oppostie direction
b1 = oS.b - Ei- oS.labelMat[i]*(oS.alphas[i]-alphaIold)*oS.K[i,i] - oS.labelMat[j]*(oS.alphas[j]-alphaJold)*oS.K[i,j]
b2 = oS.b - Ej- oS.labelMat[i]*(oS.alphas[i]-alphaIold)*oS.K[i,j]- oS.labelMat[j]*(oS.alphas[j]-alphaJold)*oS.K[j,j]
if (0 < oS.alphas[i]) and (oS.C > oS.alphas[i]): oS.b = b1
elif (0 < oS.alphas[j]) and (oS.C > oS.alphas[j]): oS.b = b2
else: oS.b = (b1 + b2)/2.0
return 1
else: return 0
def smoP(dataMatIn, classLabels, C, toler, maxIter,kTup=('lin', 0)): #full Platt SMO
oS = optStruct(mat(dataMatIn),mat(classLabels).transpose(),C,toler, kTup)
iter = 0
entireSet = True; alphaPairsChanged = 0
while (iter < maxIter) and ((alphaPairsChanged > 0) or (entireSet)):
alphaPairsChanged = 0
if entireSet: #go over all
for i in range(oS.m):
alphaPairsChanged += innerL(i,oS)
print( "fullSet, iter: %d i:%d, pairs changed %d" % (iter,i,alphaPairsChanged))
iter += 1
else:#go over non-bound (railed) alphas
nonBoundIs = nonzero((oS.alphas.A > 0) * (oS.alphas.A < C))[0]
for i in nonBoundIs:
alphaPairsChanged += innerL(i,oS)
print ("non-bound, iter: %d i:%d, pairs changed %d" % (iter,i,alphaPairsChanged))
iter += 1
if entireSet: entireSet = False #toggle entire set loop
elif (alphaPairsChanged == 0): entireSet = True
print ("iteration number: %d" % iter)
return oS.b,oS.alphas
def calcWs(alphas,dataArr,classLabels):
X = mat(dataArr); labelMat = mat(classLabels).transpose()
m,n = shape(X)
w = zeros((n,1))
for i in range(m):
w += multiply(alphas[i]*labelMat[i],X[i,:].T)
return w
def testRbf(k1=1.3):
dataArr,labelArr = loadDataSet('testSetRBF.txt')
b,alphas = smoP(dataArr, labelArr, 200, 0.0001, 10000, ('rbf', k1)) #C=200 important
datMat=mat(dataArr); labelMat = mat(labelArr).transpose()
svInd=nonzero(alphas.A>0)[0]
sVs=datMat[svInd] #get matrix of only support vectors
labelSV = labelMat[svInd];
print ("there are %d Support Vectors" % shape(sVs)[0])
m,n = shape(datMat)
errorCount = 0
for i in range(m):
kernelEval = kernelTrans(sVs,datMat[i,:],('rbf', k1))
predict=kernelEval.T * multiply(labelSV,alphas[svInd]) + b
if sign(predict)!=sign(labelArr[i]): errorCount += 1
print( "the training error rate is: %f" % (float(errorCount)/m))
dataArr,labelArr = loadDataSet('testSetRBF2.txt')
errorCount = 0
datMat=mat(dataArr); labelMat = mat(labelArr).transpose()
m,n = shape(datMat)
for i in range(m):
kernelEval = kernelTrans(sVs,datMat[i,:],('rbf', k1))
predict=kernelEval.T * multiply(labelSV,alphas[svInd]) + b
if sign(predict)!=sign(labelArr[i]): errorCount += 1
print( "the test error rate is: %f" % (float(errorCount)/m) )
def img2vector(filename):
returnVect = zeros((1,1024))
fr = open(filename)
for i in range(32):
lineStr = fr.readline()
for j in range(32):
returnVect[0,32*i+j] = int(lineStr[j])
return returnVect
def loadImages(dirName):
from os import listdir
hwLabels = []
trainingFileList = listdir(dirName) #load the training set
m = len(trainingFileList)
trainingMat = zeros((m,1024))
for i in range(m):
fileNameStr = trainingFileList[i]
fileStr = fileNameStr.split('.')[0] #take off .txt
classNumStr = int(fileStr.split('_')[0])
if classNumStr == 9: hwLabels.append(-1)
else: hwLabels.append(1)
trainingMat[i,:] = img2vector('%s/%s' % (dirName, fileNameStr))
return trainingMat, hwLabels
def testDigits(kTup=('rbf', 10)):
dataArr,labelArr = loadImages('trainingDigits')
b,alphas = smoP(dataArr, labelArr, 200, 0.0001, 10000, kTup)
datMat=mat(dataArr); labelMat = mat(labelArr).transpose()
svInd=nonzero(alphas.A>0)[0]
sVs=datMat[svInd]
labelSV = labelMat[svInd];
print( "there are %d Support Vectors" % shape(sVs)[0])
m,n = shape(datMat)
errorCount = 0
for i in range(m):
kernelEval = kernelTrans(sVs,datMat[i,:],kTup)
predict=kernelEval.T * multiply(labelSV,alphas[svInd]) + b
if sign(predict)!=sign(labelArr[i]): errorCount += 1
print ("the training error rate is: %f" % (float(errorCount)/m))
dataArr,labelArr = loadImages('testDigits')
errorCount = 0
datMat=mat(dataArr); labelMat = mat(labelArr).transpose()
m,n = shape(datMat)
for i in range(m):
kernelEval = kernelTrans(sVs,datMat[i,:],kTup)
predict=kernelEval.T * multiply(labelSV,alphas[svInd]) + b
if sign(predict)!=sign(labelArr[i]): errorCount += 1
print( "the test error rate is: %f" % (float(errorCount)/m) )
#######********************************
# Non-Kernel VErsions below
#######********************************
class optStructK:
def __init__(self,dataMatIn, classLabels, C, toler): # Initialize the structure with the parameters
self.X = dataMatIn
self.labelMat = classLabels
self.C = C
self.tol = toler
self.m = shape(dataMatIn)[0]
self.alphas = mat(zeros((self.m,1)))
self.b = 0
self.eCache = mat(zeros((self.m,2))) #first column is valid flag
def calcEkK(oS, k):
fXk = float(multiply(oS.alphas,oS.labelMat).T*(oS.X*oS.X[k,:].T)) + oS.b
Ek = fXk - float(oS.labelMat[k])
return Ek
def selectJK(i, oS, Ei): #this is the second choice -heurstic, and calcs Ej
maxK = -1; maxDeltaE = 0; Ej = 0
oS.eCache[i] = [1,Ei] #set valid #choose the alpha that gives the maximum delta E
validEcacheList = nonzero(oS.eCache[:,0].A)[0]
if (len(validEcacheList)) > 1:
for k in validEcacheList: #loop through valid Ecache values and find the one that maximizes delta E
if k == i: continue #don't calc for i, waste of time
Ek = calcEk(oS, k)
deltaE = abs(Ei - Ek)
if (deltaE > maxDeltaE):
maxK = k; maxDeltaE = deltaE; Ej = Ek
return maxK, Ej
else: #in this case (first time around) we don't have any valid eCache values
j = selectJrand(i, oS.m)
Ej = calcEk(oS, j)
return j, Ej
def updateEkK(oS, k):#after any alpha has changed update the new value in the cache
Ek = calcEk(oS, k)
oS.eCache[k] = [1,Ek]
def innerLK(i, oS):
Ei = calcEk(oS, i)
if ((oS.labelMat[i]*Ei < -oS.tol) and (oS.alphas[i] < oS.C)) or ((oS.labelMat[i]*Ei > oS.tol) and (oS.alphas[i] > 0)):
j,Ej = selectJ(i, oS, Ei) #this has been changed from selectJrand
alphaIold = oS.alphas[i].copy(); alphaJold = oS.alphas[j].copy();
if (oS.labelMat[i] != oS.labelMat[j]):
L = max(0, oS.alphas[j] - oS.alphas[i])
H = min(oS.C, oS.C + oS.alphas[j] - oS.alphas[i])
else:
L = max(0, oS.alphas[j] + oS.alphas[i] - oS.C)
H = min(oS.C, oS.alphas[j] + oS.alphas[i])
if L==H: print( "L==H"); return 0
eta = 2.0 * oS.X[i,:]*oS.X[j,:].T - oS.X[i,:]*oS.X[i,:].T - oS.X[j,:]*oS.X[j,:].T
if eta >= 0: print ("eta>=0"); return 0
oS.alphas[j] -= oS.labelMat[j]*(Ei - Ej)/eta
oS.alphas[j] = clipAlpha(oS.alphas[j],H,L)
updateEk(oS, j) #added this for the Ecache
if (abs(oS.alphas[j] - alphaJold) < 0.00001): print ("j not moving enough"); return 0
oS.alphas[i] += oS.labelMat[j]*oS.labelMat[i]*(alphaJold - oS.alphas[j])#update i by the same amount as j
updateEk(oS, i) #added this for the Ecache #the update is in the oppostie direction
b1 = oS.b - Ei- oS.labelMat[i]*(oS.alphas[i]-alphaIold)*oS.X[i,:]*oS.X[i,:].T - oS.labelMat[j]*(oS.alphas[j]-alphaJold)*oS.X[i,:]*oS.X[j,:].T
b2 = oS.b - Ej- oS.labelMat[i]*(oS.alphas[i]-alphaIold)*oS.X[i,:]*oS.X[j,:].T - oS.labelMat[j]*(oS.alphas[j]-alphaJold)*oS.X[j,:]*oS.X[j,:].T
if (0 < oS.alphas[i]) and (oS.C > oS.alphas[i]): oS.b = b1
elif (0 < oS.alphas[j]) and (oS.C > oS.alphas[j]): oS.b = b2
else: oS.b = (b1 + b2)/2.0
return 1
else: return 0
def smoPK(dataMatIn, classLabels, C, toler, maxIter): #full Platt SMO
oS = optStruct(mat(dataMatIn),mat(classLabels).transpose(),C,toler)
iter = 0
entireSet = True; alphaPairsChanged = 0
while (iter < maxIter) and ((alphaPairsChanged > 0) or (entireSet)):
alphaPairsChanged = 0
if entireSet: #go over all
for i in range(oS.m):
alphaPairsChanged += innerL(i,oS)
print ("fullSet, iter: %d i:%d, pairs changed %d" % (iter,i,alphaPairsChanged))
iter += 1
else:#go over non-bound (railed) alphas
nonBoundIs = nonzero((oS.alphas.A > 0) * (oS.alphas.A < C))[0]
for i in nonBoundIs:
alphaPairsChanged += innerL(i,oS)
print ("non-bound, iter: %d i:%d, pairs changed %d" % (iter,i,alphaPairsChanged))
iter += 1
if entireSet: entireSet = False #toggle entire set loop
elif (alphaPairsChanged == 0): entireSet = True
print ("iteration number: %d" % iter)
return oS.b,oS.alphas
绘画数据点的draw.py文件:
# -*- coding: utf-8 -*-
"""
Created on Sun May 6 00:34:51 2018
@author: Diky
"""
import matplotlib
import matplotlib.pyplot as plt
import numpy as np
def showDataSet(dataMat, labelMat):
data_plus = [] #正样本
data_minus = [] #负样本
for i in range(len(dataMat)):
if labelMat[i] > 0:
data_plus.append(dataMat[i])
else:
data_minus.append(dataMat[i])
data_plus_np = np.array(data_plus) #转换为numpy矩阵
data_minus_np = np.array(data_minus) #转换为numpy矩阵
plt.scatter(np.transpose(data_plus_np)[0], np.transpose(data_plus_np)[1]) #正样本散点图
plt.scatter(np.transpose(data_minus_np)[0], np.transpose(data_minus_np)[1]) #负样本散点图
plt.show()
def showClassifer(dataMat,labelMat , w, b ,alphas):
#绘制样本点
data_plus = [] #正样本
data_minus = [] #负样本
for i in range(len(dataMat)):
if labelMat[i] > 0:
data_plus.append(dataMat[i])
else:
data_minus.append(dataMat[i])
data_plus_np = np.array(data_plus) #转换为numpy矩阵
data_minus_np = np.array(data_minus) #转换为numpy矩阵
plt.scatter(np.transpose(data_plus_np)[0], np.transpose(data_plus_np)[1], s=30, alpha=0.7) #正样本散点图
plt.scatter(np.transpose(data_minus_np)[0], np.transpose(data_minus_np)[1], s=30, alpha=0.7) #负样本散点图
#绘制直线
x1 = max(dataMat)[0]
x2 = min(dataMat)[0]
a1, a2 = w
b = float(b)
a1 = float(a1[0])
a2 = float(a2[0])
y1, y2 = (-b- a1*x1)/a2, (-b - a1*x2)/a2
plt.plot([x1, x2], [y1, y2])
#找出支持向量点
for i, alpha in enumerate(alphas):
if abs(alpha) > 0:
x, y = dataMat[i]
plt.scatter([x], [y], s=150, c='none', alpha=0.7, linewidth=1.5, edgecolor='red')
plt.show()
def get_w(dataMat, labelMat, alphas):
alphas, dataMat, labelMat = np.array(alphas), np.array(dataMat), np.array(labelMat)
w = np.dot((np.tile(labelMat.reshape(1, -1).T, (1, 2)) * dataMat).T, alphas)
return w.tolist()
测试主函数main.py为:
# -*- coding: utf-8 -*-
"""
Created on Sat May 5 23:56:01 2018
@author: Diky
"""
from svmMLiA import *
from draw import *
dataMat, labelMat = loadDataSet('testSet.txt')
b1,alphas1 = smoSimple(dataMat, labelMat, 0.6, 0.001, 40)
ws1=calcWs(alphas1,dataArr,labelArr)
b2,alphas2 = smoP(dataMat, labelMat, 0.6, 0.001, 40)
ws2=calcWs(alphas2,dataArr,labelArr)
print('\n\ninitial:')
showDataSet(dataMat, labelMat)
print('SVM simple :')
showClassifer(dataMat, labelMat ,ws1, b1 ,alphas1)
print('SVM completed :')
showClassifer(dataMat, labelMat ,ws2, b2 ,alphas2)
testRbf()
dataMat, labelMat = loadDataSet('testSetRBF.txt')
showDataSet(dataMat, labelMat)
#testDigits(('lin',10)) # ' lin ' and' rbf'
testDigits(kTup=('rbf', 100))