解析pubmed文献数据库中的xml文章结构-01
一、下载文章,查看文章结构
相信大部分做生物医学领域的,对pubmed文献数据库都比较熟悉,今天主要是利用pubmed提供的文献检索数据库,下载对应的xml文章结构,利用python对其进行解析,导入mysql数据库中。
首先,pubmed文献数据库的网址是https://www.ncbi.nlm.nih.gov/pubmed/ ,下载了部分年份的文章,文章结构主要如下:
<PubmedArticle>
<MedlineCitation Status="MEDLINE" Owner="NLM">
<PMID Version="1">25534978PMID>
<DateCompleted>
<Year>2015Year>
<Month>08Month>
<Day>21Day>
DateCompleted>
<DateRevised>
<Year>2016Year>
<Month>12Month>
<Day>15Day>
DateRevised>
<Article PubModel="Print">
<Journal>
<ISSN IssnType="Electronic">1744-8409ISSN>
<JournalIssue CitedMedium="Internet">
<Volume>11Volume>
<Issue>1Issue>
<PubDate>
<Year>2015Year>
<Month>JanMonth>
PubDate>
JournalIssue>
<Title>Expert review of clinical immunologyTitle>
<ISOAbbreviation>Expert Rev Clin ImmunolISOAbbreviation>
Journal>
<ArticleTitle>Autoimmune disease in the epigenetic era: how has epigenetics changed our understanding of disease and how can we expect the field to evolve?ArticleTitle>
<Pagination>
<MedlinePgn>45-58MedlinePgn>
Pagination>
<ELocationID EIdType="doi" ValidYN="Y">10.1586/1744666X.2015.994507ELocationID>
<Abstract>
<AbstractText>Autoimmune diseases are complex and enigmatic, and have presented particular challenges to researchers seeking to define their etiology and explain progression. Previous studies have implicated epigenetic influences in the development of autoimmunity. Epigenetics describes changes in gene expression related to environmental influences without alterations in the underlying genomic sequence, generally classified into three main groups: cytosine genomic DNA methylation, modification of various sidechain positions of histone proteins and noncoding RNAs feedback. The purpose of this article is to review the most relevant literature describing alterations of epigenetic marks in the development and progression of four common autoimmune diseases: systemic lupus erythematosus, rheumatoid arthritis, systemic sclerosis and Sjögren's syndrome. The contribution of DNA methylation, histone modification and noncoding RNA for each of these disorders is discussed, including examples both of candidate gene studies and larger epigenomics surveys, and in various tissue types important for the pathogenesis of each. The future of the field is speculated briefly, as is the possibility of therapeutic interventions targeting the epigenome. AbstractText>
Abstract>
<AuthorList CompleteYN="Y">
<Author ValidYN="Y">
<LastName>JeffriesLastName>
<ForeName>Matlock AForeName>
<Initials>MAInitials>
<AffiliationInfo>
<Affiliation>Department of Internal Medicine, Division of Rheumatology, Immunology and Allergy, University of Oklahoma Health Sciences Center, Oklahoma City, OK, USA.Affiliation>
AffiliationInfo>
Author>
<Author ValidYN="Y">
<LastName>SawalhaLastName>
<ForeName>Amr HForeName>
<Initials>AHInitials>
Author>
AuthorList>
<Language>engLanguage>
<GrantList CompleteYN="Y">
<Grant>
<GrantID>R01 AI097134GrantID>
<Acronym>AIAcronym>
<Agency>NIAID NIH HHSAgency>
<Country>United StatesCountry>
Grant>
<Grant>
<GrantID>R01AI097134GrantID>
<Acronym>AIAcronym>
<Agency>NIAID NIH HHSAgency>
<Country>United StatesCountry>
Grant>
GrantList>
<PublicationTypeList>
<PublicationType UI="D016428">Journal ArticlePublicationType>
<PublicationType UI="D052061">Research Support, N.I.H., ExtramuralPublicationType>
<PublicationType UI="D016454">ReviewPublicationType>
PublicationTypeList>
Article>
<MedlineJournalInfo>
<Country>EnglandCountry>
<MedlineTA>Expert Rev Clin ImmunolMedlineTA>
<NlmUniqueID>101271248NlmUniqueID>
<ISSNLinking>1744-666XISSNLinking>
MedlineJournalInfo>
<ChemicalList>
<Chemical>
<RegistryNumber>0RegistryNumber>
<NameOfSubstance UI="D006657">HistonesNameOfSubstance>
Chemical>
<Chemical>
<RegistryNumber>0RegistryNumber>
<NameOfSubstance UI="D022661">RNA, UntranslatedNameOfSubstance>
Chemical>
ChemicalList>
<CitationSubset>IMCitationSubset>
<CommentsCorrectionsList>
<CommentsCorrections RefType="Cites">
<RefSource>Eur J Immunol. 2007 May;37(5):1407-13RefSource>
<PMID Version="1">17429846PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Nature. 2007 May 24;447(7143):396-8RefSource>
<PMID Version="1">17522671PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Arthritis Rheum. 2011 May;63(5):1376-86RefSource>
<PMID Version="1">21538319PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Arthritis Rheum. 2011 May;63(5):1452-8RefSource>
<PMID Version="1">21538322PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Epigenetics. 2011 May;6(5):593-601RefSource>
<PMID Version="1">21436623PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Eur J Immunol. 2011 Jul;41(7):2029-39RefSource>
<PMID Version="1">21469088PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Blood. 2011 Aug 11;118(6):1472-80RefSource>
<PMID Version="1">21613261PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Cell Cycle. 2011 Aug 15;10(16):2662-8RefSource>
<PMID Version="1">21811096PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Genes Immun. 2011 Dec;12(8):643-52RefSource>
<PMID Version="1">21753787PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>J Transl Med. 2011;9:192RefSource>
<PMID Version="1">22060015PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>PLoS One. 2011;6(11):e28104RefSource>
<PMID Version="1">22140515PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Clin Immunol. 2012 Apr;143(1):39-44RefSource>
<PMID Version="1">22306512PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>J Immunol. 2010 Mar 1;184(5):2718-28RefSource>
<PMID Version="1">20100935PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Genes Immun. 2010 Mar;11(2):124-33RefSource>
<PMID Version="1">19710693PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Arthritis Rheum. 2010 May;62(5):1438-47RefSource>
<PMID Version="1">20131288PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>J Autoimmun. 2010 Aug;35(1):58-69RefSource>
<PMID Version="1">20223637PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>J Immunol. 2010 Jun 15;184(12):6773-81RefSource>
<PMID Version="1">20483747PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Arthritis Rheum. 2010 Jun;62(6):1733-43RefSource>
<PMID Version="1">20201077PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Clin Rev Allergy Immunol. 2010 Aug;39(1):78-84RefSource>
<PMID Version="1">19662539PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>J Biomed Biotechnol. 2010;2010:931018RefSource>
<PMID Version="1">20589076PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Arthritis Res Ther. 2010;12(3):R81RefSource>
<PMID Version="1">20459811PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Cancer. 2010 Sep 1;116(17):4043-53RefSource>
<PMID Version="1">20564122PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>J Dermatol Sci. 2010 Sep;59(3):198-203RefSource>
<PMID Version="1">20724115PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>J Biol Chem. 2013 Jul 26;288(30):21936-44RefSource>
<PMID Version="1">23775084PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>PLoS Genet. 2013;9(8):e1003678RefSource>
<PMID Version="1">23950730PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Clin Immunol. 2013 Oct;149(1):46-54RefSource>
<PMID Version="1">23891737PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>J Immunol. 2012 Apr 15;188(8):3567-71RefSource>
<PMID Version="1">22422882PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Ann Rheum Dis. 2000 Jun;59(6):455-61RefSource>
<PMID Version="1">10834863PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Arthritis Rheum. 2000 Dec;43(12):2634-47RefSource>
<PMID Version="1">11145021PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Arthritis Rheum. 2000 Dec;43(12):2807-17RefSource>
<PMID Version="1">11145040PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Arthritis Rheum. 2002 May;46(5):1282-91RefSource>
<PMID Version="1">12115234PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Arthritis Rheum. 2012 Jul;64(7):2338-45RefSource>
<PMID Version="1">22231486PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Genes Immun. 2012 Jul;13(5):388-98RefSource>
<PMID Version="1">22495533PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Rheumatology (Oxford). 2012 Sep;51(9):1550-6RefSource>
<PMID Version="1">22661558PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Arthritis Rheum. 2012 Sep;64(9):2964-74RefSource>
<PMID Version="1">22549474PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Clin Immunol. 2012 Oct;145(1):13-8RefSource>
<PMID Version="1">22889643PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Ann Rheum Dis. 2013 Jan;72(1):110-7RefSource>
<PMID Version="1">22736089PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>J Immunol. 2013 Feb 1;190(3):1297-303RefSource>
<PMID Version="1">23277489PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Arthritis Rheum. 2013 Feb;65(2):481-91RefSource>
<PMID Version="1">23045159PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Nat Genet. 2013 Feb;45(2):124-30RefSource>
<PMID Version="1">23263488PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Nat Biotechnol. 2013 Feb;31(2):142-7RefSource>
<PMID Version="1">23334450PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Ann Rheum Dis. 2013 Apr;72(4):614-20RefSource>
<PMID Version="1">22915621PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>J Autoimmun. 2013 Mar;41:6-16RefSource>
<PMID Version="1">23306098PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>J Autoimmun. 2013 Mar;41:168-74RefSource>
<PMID Version="1">23428850PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>J Autoimmun. 2013 Mar;41:175-81RefSource>
<PMID Version="1">23478041PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>J Autoimmun. 2013 Jun;43:78-84RefSource>
<PMID Version="1">23623029PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>J Clin Immunol. 2013 Aug;33(6):1100-9RefSource>
<PMID Version="1">23657402PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Clin Immunol. 2013 Aug;148(2):254-7RefSource>
<PMID Version="1">23773924PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>FEBS Lett. 2014 Nov 17;588(22):4244-9RefSource>
<PMID Version="1">24873878PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Curr Opin Immunol. 2014 Dec;31:16-23RefSource>
<PMID Version="1">25214301PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Ann Rheum Dis. 2015 Jun;74(6):1265-74RefSource>
<PMID Version="1">24562503PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Ann Rheum Dis. 2015 Aug;74(8):1612-20RefSource>
<PMID Version="1">24812288PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Annu Rev Immunol. 2005;23:307-36RefSource>
<PMID Version="1">15771573PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>J Biol Chem. 2005 Dec 9;280(49):40749-56RefSource>
<PMID Version="1">16230360PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>J Proteome Res. 2005 Nov-Dec;4(6):2032-42RefSource>
<PMID Version="1">16335948PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Curr Dir Autoimmun. 2006;9:173-87RefSource>
<PMID Version="1">16394661PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Arthritis Res Ther. 2010;12(4):R133RefSource>
<PMID Version="1">20609223PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Mol Biotechnol. 2010 Nov;46(3):243-9RefSource>
<PMID Version="1">20563671PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Mod Rheumatol. 2010 Oct;20(5):458-65RefSource>
<PMID Version="1">20490598PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Genes Immun. 2010 Oct;11(7):554-60RefSource>
<PMID Version="1">20463746PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Rheumatol Int. 2010 Nov;30(12):1627-33RefSource>
<PMID Version="1">20049450PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>J Immunol. 2010 Nov 15;185(10):6355-63RefSource>
<PMID Version="1">20952683PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Arthritis Rheum. 2012 Jun;64(6):1809-17RefSource>
<PMID Version="1">22170508PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Mol Ther. 2012 Jun;20(6):1251-60RefSource>
<PMID Version="1">22395530PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>J Biol Chem. 2003 Feb 14;278(7):4806-12RefSource>
<PMID Version="1">12473678PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Eur J Immunol. 2003 Oct;33(10):2792-800RefSource>
<PMID Version="1">14515263PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>J Immunol. 2004 Mar 15;172(6):3652-61RefSource>
<PMID Version="1">15004168PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Autoimmunity. 2004 Feb;37(1):57-65RefSource>
<PMID Version="1">15115313PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Arthritis Rheum. 2004 Jun;50(6):1850-60RefSource>
<PMID Version="1">15188362PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Arthritis Rheum. 2004 Oct;50(10):3365-76RefSource>
<PMID Version="1">15476220PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Proc Natl Acad Sci U S A. 1967 May;57(5):1394-400RefSource>
<PMID Version="1">5231746PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Proc Natl Acad Sci U S A. 1985 Dec;82(24):8629-33RefSource>
<PMID Version="1">2417226PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Hum Immunol. 1986 Dec;17(4):456-70RefSource>
<PMID Version="1">2432050PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Arthritis Rheum. 1990 Nov;33(11):1665-73RefSource>
<PMID Version="1">2242063PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>J Rheumatol. 1991 Apr;18(4):530-4RefSource>
<PMID Version="1">2066944PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>J Immunol. 1991 Sep 1;147(5):1477-83RefSource>
<PMID Version="1">1715359PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>J Chromatogr. 1991 May 31;566(2):481-91RefSource>
<PMID Version="1">1939459PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>J Clin Invest. 1993 Jul;92(1):38-53RefSource>
<PMID Version="1">7686923PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>J Immunol. 1995 Feb 1;154(3):1470-80RefSource>
<PMID Version="1">7529804PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>J Immunol. 1995 Mar 15;154(6):3025-35RefSource>
<PMID Version="1">7533191PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Lupus. 1997;6(3):326-7RefSource>
<PMID Version="1">9296780PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Nat Genet. 1998 Jun;19(2):187-91RefSource>
<PMID Version="1">9620779PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Arthritis Rheum. 2005 Jan;52(1):201-11RefSource>
<PMID Version="1">15641052PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Ann Rheum Dis. 2005 Mar;64(3):481-3RefSource>
<PMID Version="1">15708899PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Epigenetics. 2013 Jul;8(7):679-84RefSource>
<PMID Version="1">23803967PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Autoimmun Rev. 2013 Oct;12(12):1160-5RefSource>
<PMID Version="1">23860189PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Nat Rev Rheumatol. 2013 Nov;9(11):674-86RefSource>
<PMID Version="1">24100461PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Nat Biotechnol. 2013 Dec;31(12):1137-42RefSource>
<PMID Version="1">24108092PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Arthritis Rheumatol. 2014 Mar;66(3):549-59RefSource>
<PMID Version="1">24574214PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Ann Rheum Dis. 2014 Jun;73(6):1232-9RefSource>
<PMID Version="1">23698475PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Acta Histochem. 2014 Jun;116(5):891-7RefSource>
<PMID Version="1">24657071PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Clin Exp Immunol. 2014 Sep;177(3):641-51RefSource>
<PMID Version="1">24816316PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Genome Biol. 2013;14(3):R21RefSource>
<PMID Version="1">23497655PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Genome Biol. 2014;15(2):R31RefSource>
<PMID Version="1">24495553PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Arthritis Rheumatol. 2014 Oct;66(10):2804-15RefSource>
<PMID Version="1">24980887PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Nat Commun. 2012;3:735RefSource>
<PMID Version="1">22415826PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>J Immunol. 2012 Apr 1;188(7):3323-31RefSource>
<PMID Version="1">22379029PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Arthritis Rheum. 2007 Jun;56(6):1921-33RefSource>
<PMID Version="1">17530637PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Arthritis Rheum. 2007 Aug;56(8):2755-64RefSource>
<PMID Version="1">17665426PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>J Immunol. 2007 Oct 15;179(8):5553-63RefSource>
<PMID Version="1">17911642PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>J Immunol. 2007 Nov 1;179(9):6352-8RefSource>
<PMID Version="1">17947713PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Rheumatology (Oxford). 2007 Dec;46(12):1796-803RefSource>
<PMID Version="1">18032537PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Ann Rheum Dis. 2008 Jun;67(6):867-72RefSource>
<PMID Version="1">17823201PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Nat Genet. 2008 Jun;40(6):741-50RefSource>
<PMID Version="1">18488029PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Genes Immun. 2008 Jun;9(4):368-78RefSource>
<PMID Version="1">18523434PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Arthritis Rheum. 2008 Aug;58(8):2511-7RefSource>
<PMID Version="1">18668569PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Arthritis Rheum. 2008 Nov;58(11):3562-73RefSource>
<PMID Version="1">18975310PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Arthritis Rheum. 2009 May;60(5):1519-29RefSource>
<PMID Version="1">19404935PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>J Biol Chem. 2009 Jul 3;284(27):17897-901RefSource>
<PMID Version="1">19342379PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Clin Immunol. 2009 Sep;132(3):362-70RefSource>
<PMID Version="1">19520616PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>J Rheumatol. 2009 Aug;36(8):1580-9RefSource>
<PMID Version="1">19531758PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>J Immunol. 2009 Sep 1;183(5):3109-17RefSource>
<PMID Version="1">19648272PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>PLoS One. 2009;4(8):e6718RefSource>
<PMID Version="1">19701459PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>ISME J. 2011 Jan;5(1):82-91RefSource>
<PMID Version="1">20613793PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Immunol Lett. 2011 Mar 30;135(1-2):96-9RefSource>
<PMID Version="1">20937307PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>J Dermatol Sci. 2009 Oct;56(1):33-6RefSource>
<PMID Version="1">19651491PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Lupus. 2009 Oct;18(12):1037-44RefSource>
<PMID Version="1">19762376PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Nat Genet. 2009 Nov;41(11):1228-33RefSource>
<PMID Version="1">19838195PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Nature. 2009 Nov 19;462(7271):315-22RefSource>
<PMID Version="1">19829295PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Mol Immunol. 2009 Dec;47(2-3):511-6RefSource>
<PMID Version="1">19747733PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Arthritis Rheum. 2009 Dec;60(12):3613-22RefSource>
<PMID Version="1">19950268PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Genome Res. 2010 Feb;20(2):170-9RefSource>
<PMID Version="1">20028698PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Scand J Rheumatol. 2009;38(5):369-74RefSource>
<PMID Version="1">19444718PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Arthritis Rheum. 2006 Mar;54(3):779-87RefSource>
<PMID Version="1">16508942PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>J Autoimmun. 2006 May;26(3):165-71RefSource>
<PMID Version="1">16621447PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Arthritis Rheum. 2006 Jul;54(7):2271-9RefSource>
<PMID Version="1">16802366PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Br J Pharmacol. 2007 Apr;150(7):862-72RefSource>
<PMID Version="1">17325656PMID>
CommentsCorrections>
<CommentsCorrections RefType="Cites">
<RefSource>Clin Rheumatol. 2007 May;26(5):723-8RefSource>
<PMID Version="1">17103120PMID>
CommentsCorrections>
CommentsCorrectionsList>
<MeshHeadingList>
<MeshHeading>
<DescriptorName UI="D000818" MajorTopicYN="N">AnimalsDescriptorName>
MeshHeading>
<MeshHeading>
<DescriptorName UI="D001327" MajorTopicYN="Y">Autoimmune DiseasesDescriptorName>
<QualifierName UI="Q000235" MajorTopicYN="N">geneticsQualifierName>
<QualifierName UI="Q000276" MajorTopicYN="N">immunologyQualifierName>
MeshHeading>
<MeshHeading>
<DescriptorName UI="D019175" MajorTopicYN="N">DNA MethylationDescriptorName>
<QualifierName UI="Q000276" MajorTopicYN="Y">immunologyQualifierName>
MeshHeading>
<MeshHeading>
<DescriptorName UI="D044127" MajorTopicYN="N">Epigenesis, GeneticDescriptorName>
<QualifierName UI="Q000276" MajorTopicYN="Y">immunologyQualifierName>
MeshHeading>
<MeshHeading>
<DescriptorName UI="D059647" MajorTopicYN="Y">Gene-Environment InteractionDescriptorName>
MeshHeading>
<MeshHeading>
<DescriptorName UI="D006657" MajorTopicYN="Y">HistonesDescriptorName>
<QualifierName UI="Q000235" MajorTopicYN="N">geneticsQualifierName>
<QualifierName UI="Q000276" MajorTopicYN="N">immunologyQualifierName>
MeshHeading>
<MeshHeading>
<DescriptorName UI="D006801" MajorTopicYN="N">HumansDescriptorName>
MeshHeading>
<MeshHeading>
<DescriptorName UI="D011499" MajorTopicYN="N">Protein Processing, Post-TranslationalDescriptorName>
<QualifierName UI="Q000235" MajorTopicYN="N">geneticsQualifierName>
<QualifierName UI="Q000276" MajorTopicYN="Y">immunologyQualifierName>
MeshHeading>
<MeshHeading>
<DescriptorName UI="D022661" MajorTopicYN="Y">RNA, UntranslatedDescriptorName>
<QualifierName UI="Q000235" MajorTopicYN="N">geneticsQualifierName>
<QualifierName UI="Q000276" MajorTopicYN="N">immunologyQualifierName>
MeshHeading>
MeshHeadingList>
<KeywordList Owner="NOTNLM">
<Keyword MajorTopicYN="N">Sjögren’s syndromeKeyword>
<Keyword MajorTopicYN="N">autoimmune diseaseKeyword>
<Keyword MajorTopicYN="N">epigeneticsKeyword>
<Keyword MajorTopicYN="N">histone modificationKeyword>
<Keyword MajorTopicYN="N">methylationKeyword>
<Keyword MajorTopicYN="N">miRNAKeyword>
<Keyword MajorTopicYN="N">rheumatoid arthritisKeyword>
<Keyword MajorTopicYN="N">systemic lupus erythematosusKeyword>
<Keyword MajorTopicYN="N">systemic sclerosisKeyword>
KeywordList>
MedlineCitation>
<PubmedData>
<History>
<PubMedPubDate PubStatus="entrez">
<Year>2014Year>
<Month>12Month>
<Day>24Day>
<Hour>6Hour>
<Minute>0Minute>
PubMedPubDate>
<PubMedPubDate PubStatus="pubmed">
<Year>2014Year>
<Month>12Month>
<Day>24Day>
<Hour>6Hour>
<Minute>0Minute>
PubMedPubDate>
<PubMedPubDate PubStatus="medline">
<Year>2015Year>
<Month>8Month>
<Day>22Day>
<Hour>6Hour>
<Minute>0Minute>
PubMedPubDate>
History>
<PublicationStatus>ppublishPublicationStatus>
<ArticleIdList>
<ArticleId IdType="pubmed">25534978ArticleId>
<ArticleId IdType="doi">10.1586/1744666X.2015.994507ArticleId>
<ArticleId IdType="pmc">PMC4636192ArticleId>
<ArticleId IdType="mid">NIHMS732942ArticleId>
ArticleIdList>
PubmedData>
PubmedArticle>由于文章结构嵌套了很多的节点信息,要提取有用的信息,需要进行层次化的提取,主要采用了python中的xml.ElementTree的语法对其进解析。代码如下:
"""
@Author: ly
@Date: 2018-07-30 10:31:16
import xml.etree.ElementTree as ET
import pandas as pd
import numpy as np
# 国家信息
# 存在问题:有的作者单位中的国家名称不统一,例如:美国(USA, UNITED STATES)
# 有的作者单位中没国家,只写了州
# 目前解决方法:通过查看缺失或无法识别国家信息的xml文档,添加可以识别国家信息的词到词库
import pycountry
country_name = [
str.strip(str.split(i.name.upper(), ",")[0])
for i in list(pycountry.countries)
]
# 美国和英国有多种表示方式
# 添加已找到的一些国家或省份信息
country_name.extend([
"USA", "UK.", "UK ", "LONDON", "São Paulo", "IRAN", "México", "Birmingham",
"Chicago", "Deutschland", "Tokyo", "Nagoya ", "España", "serbia", "paris",
"pennsylvania", "birmingham", "chicago", "nagoya", "España",
"Belo Horizonte", "CHINESE","San Pietro Vernotico"
])
country_name = [i.upper() for i in country_name]
# 整合用于识别同一个国家的多种代表性词
# 例如:整合USA, UNITED STATES 为 USA
def CombineCountry(CountryInfo):
usa = [
"USA", "UNITED STATES", "CHICAGO", "BIRMINGHAM", "PENNSYLVANIA",
"BIRMINGHAM"
]
uk = ["UK.", "UK ", "UNITED KINGDOM", "LONDON"]
germany = ["GERMANY", "DEUTSCHLAND"]
mexico = ["MÉXICO", "MEXICO"]
japan = ["JAPAN", "TOKYO", "NAGOYA"]
barzil = ["BRAZIL", "SÃO PAULO", "BELO HORIZONTE"]
france = ["FRANCE", "PARIS"]
spain = ["SPAIN", "ESPAÑA"]
china = ["CHINA", "HONG KONG", "MACAO", "CHINESE", "TAIWAN"]
italy=["ITALY","SAN PIETRO VERNOTICO"]
CountryInfo_arr = []
for i in np.arange(len(CountryInfo)):
if CountryInfo[i] in usa:
CountryInfo_arr.append("USA")
continue
elif CountryInfo[i] in uk:
CountryInfo_arr.append("UK")
continue
elif CountryInfo[i] in germany:
CountryInfo_arr.append("GERMANY")
continue
elif CountryInfo[i] in mexico:
CountryInfo_arr.append("MEXICO")
continue
elif CountryInfo[i] in japan:
CountryInfo_arr.append("JAPAN")
continue
elif CountryInfo[i] in barzil:
CountryInfo_arr.append("BRAZIL")
continue
elif CountryInfo[i] in france:
CountryInfo_arr.append("FRANCE")
continue
elif CountryInfo[i] in spain:
CountryInfo_arr.append("SPAIN")
continue
elif CountryInfo[i] in china:
CountryInfo_arr.append("CHINA")
continue
elif CountryInfo[i] in italy:
CountryInfo_arr.append("ITALY")
continue
CountryInfo_arr.append(CountryInfo[i])
return (CountryInfo_arr)
# 用于识别单位
def IdentifyInstitute(authorAff):
# 用于识别机构的词库,有的无法识别时需要更新词库
org=["UNIVERSITY","COMPANY","INSTITUTET","COLLEGE","ACADEMY"]
# 用“,”分割单位信息,再用org词库去识别机构
string_list=[str.strip(i.upper()) for i in str.split(authorAff,",")]
author_institutet=""
for i in string_list:
for j in org:
if j in i:
author_institutet=i
break
if author_institutet!="":
break
return(author_institutet)
# 识别一作的国别,机构
def FirstAuthorCountry(Affiliation,country_name):
firstAuthorCountry = []
firstAuthorInstitute=[]
# 提取第一作者单位信息
# count = 0
for i in Affiliation:
# count += 1
# print(count)
firstAuthorCountry_temp = ""
firstAuthorInstitute_temp=""
if i != [] and i[0] != []:
# 一作单位信息
firstAuthorAff = i[0][0].upper()
firstAuthorInstitute_temp=IdentifyInstitute(firstAuthorAff)
# 匹配国家
for j in country_name:
if j in firstAuthorAff:
firstAuthorCountry_temp = j
break
firstAuthorCountry.append(firstAuthorCountry_temp)
firstAuthorCountry=CombineCountry(firstAuthorCountry)
firstAuthorInstitute.append(firstAuthorInstitute_temp)
return ([firstAuthorCountry,firstAuthorInstitute])
# 根据email地址识别通讯作者
# 匹配邮箱地址,如果多个作者单位信息包含邮箱地址,则返回最后一位,
# 如果没有作者单位信息包含邮箱地址,也返回最后一位作者为通讯作者
import re
def IdentifyContactIndex(Affiliation,country_name):
# 匹配邮箱地址正则表达式
pattern = re.compile(r'\S+@\S+')
# 保存通讯作者的下标
contect_index_arr = []
# count = 0
for i in Affiliation:
# count += 1
# print(count, "\n")
# 每篇论文所有作者单位信息
contect_index_temp = []
author_index = 0
# flag=1表示存在作者单位包含邮箱,如果都不包含邮箱,则最后一位作者为通讯作者
flag = 0
# 每个作者单位信息
for j in i:
# 有的作者挂了多个单位,如果单位地址包含邮箱则返回作者下标
for k in j:
if len(pattern.findall(k)) > 0:
contect_index_temp.append(author_index)
flag = 1
break
author_index += 1
if flag == 0:
contect_index_arr.append(len(i) - 1)
else:
# 存在多个作者有邮箱地址,取最后一位作者为通讯作者
contect_index_arr.append(contect_index_temp[-1])
return (contect_index_arr)
# 通讯作者国别
def ContectAuthorCountry(Affiliation,country_name):
contectAuthorIndex = IdentifyContactIndex(Affiliation,country_name)
# # 取存在邮箱地址最后一位作者为通讯作者,一般都是最后一位为通讯作者
# connAuthorIndex=[i[-1] for i in connAuthorIndex]
contectAuthorCountry = []
contectAuthorInstitute=[]
# 提取第一作者单位信息
# count = 0
for (i, j) in zip(Affiliation, contectAuthorIndex):
# count += 1
# print(count)
contectAuthorCountry_temp = ""
contectAuthorInstitute_temp=""
if i != [] and i[j] != []:
contectAuthorAff = i[j][0].upper()
contectAuthorInstitute_temp=IdentifyInstitute(contectAuthorAff)
# 匹配国家
for k in country_name:
if k in contectAuthorAff:
contectAuthorCountry_temp = k
break
contectAuthorCountry.append(contectAuthorCountry_temp)
contectAuthorCountry=CombineCountry(contectAuthorCountry)
contectAuthorInstitute.append(contectAuthorInstitute_temp)
return ([contectAuthorCountry,contectAuthorInstitute])
# 识别每个作者的国别,用于生成合作关系图
def EachAuthorCountry(Affiliation,country_name):
authorCountry_arr = []
# count = 0
for i in Affiliation:
# count += 1
# print(count)
authorCountry = []
if i != []:
# 遍历作者单位信息
for j in i:
if j==[]:
continue
authorAff = j[0].upper()
# 匹配国家
authorCountry_temp = ""
for k in country_name:
if k in authorAff:
authorCountry_temp = k
break
if authorCountry_temp!="":
authorCountry.append(authorCountry_temp)
authorCountry_arr.append(authorCountry)
# 国家名规范化
authorCountry_std = []
for country in authorCountry_arr:
authorCountry_std.append(CombineCountry(country))
return (authorCountry_std)
# 作者国别合作连线
from itertools import combinations
# def CountryLink(Affiliation,country_name):
# authorCountry=EachAuthorCountry(Affiliation,country_name)
# countryLink = []
# for i in authorCountry:
# if len(np.unique(i)) > 1:
# countryLink.extend(list(combinations(np.unique(i), r=2)))
# countryLink=[[i[0],i[1]] for i in countryLink]
# countryLink=pd.DataFrame(countryLink)
# # countryLink.to_csv("countryLink.csv")
# return (countryLink)
def CountryLink(EachAuthorCountryInfo):
countryLink = []
for i in EachAuthorCountryInfo:
if len(np.unique(i)) > 1:
countryLink.extend(list(combinations(np.unique(i), r=2)))
countryLink=[[i[0],i[1]] for i in countryLink]
countryLink=pd.DataFrame(countryLink)
countryLink.to_csv("CountryLink.csv")
return (countryLink)
# 论文的所有作者的国别拼接成字符串
def read_xml(path):
tree = ET.parse(path)
all = tree.findall("./")
book = tree.findall("PubmedBookArticle")
art = tree.findall("PubmedArticle")
print("the numbers of article: ", len(art), "\n")
print("the numbers of book: ", len(book), "\n")
print("the numbers of all iterms: ", len(all), "\n")
# 2018影响因子
if2018=pd.read_csv("IF2018.csv").values
pmid_arr = []
articleTitle_arr = []
articleAbstract_arr = []
pubData_arr = []
MESH_majorTerms_arr = []
MESH_allTerms_arr = []
jornalName_arr = []
jornalNameAbbr_arr = []
authorNameList_arr = []
authorAff_arr = []
citedList_arr = []
grantInfoList_arr = []
if2018_arr=[]
count = 0
for paper in art:
pmid = "None"
pubData = "None"
MESH_majorTerms = []
MESH_allTerms = []
articleTitle = "None"
articleAbstract = "None"
jornalName = "None"
jornalNameAbbr = "None"
authorNameList = []
citedList = []
authorAff = []
temp = paper.find("MedlineCitation")
pmid = temp.find("PMID").text
print(pmid)
# RetractionOf=1时,文章被撤稿
RetractionOf = 0
# pubData 只保存了发表年,pubmed xml 文件中pubData有的只有年,有的只有年月,有的有年月日
if temp.find("Article").find("Journal").find("JournalIssue").find(
"PubDate") != None:
if temp.find("Article").find("Journal").find("JournalIssue").find(
"PubDate").find("Year") != None:
pubData = temp.find("Article").find("Journal").find(
"JournalIssue").find("PubDate").find("Year").text
elif temp.find("Article").find("Journal").find(
"JournalIssue").find("PubDate").find(
"MedlineDate") != None:
# The date of publication of the article will be found in when parsing for the separate fields is not possible.
# i.e.,1998 Dec-1999 Jan , 2000 Spring
# from url: https://www.nlm.nih.gov/bsd/licensee/elements_descriptions.html#pubdate
pubData = str.split(
temp.find("Article").find("Journal").find("JournalIssue")
.find("PubDate").find("MedlineDate").text, " ")[0]
jornalName = temp.find("Article").find("Journal").find("Title").text
# article title
# pmid: 26623013
# Tripterygium glycosides inhibit inflammatory mediators in the rat synovial RSC-364 cell line stimulated with interleukin-1β.
if temp.find("Article").find("ArticleTitle") != None:
if temp.find("Article").find("ArticleTitle").find("i") != None:
articleTitle = temp.find("Article").find("ArticleTitle").find(
"i").text + temp.find("Article").find("ArticleTitle").find(
"i").tail
else:
articleTitle = temp.find("Article").find("ArticleTitle").text
# article abstract
# pmid: 26623013
# Tripterygium glycosides *****
if temp.find("Article").find("Abstract") != None:
for i in temp.find("Article").find("Abstract").findall(
"AbstractText"):
if i.findall("./")!=[]:
for j in i.findall("./"):
if j.text !=None:
articleAbstract+=" "+j.text
if j.tail!=None:
articleAbstract+=j.tail
elif i.text!=None:
articleAbstract += " " + i.text
articleAbstract = str.strip(articleAbstract)
# 期刊名缩写,没有ISOAbbreviation时就用Title
if temp.find("Article").find("Journal").find(
"ISOAbbreviation") != None:
jornalNameAbbr = temp.find("Article").find("Journal").find(
"ISOAbbreviation").text
else:
jornalNameAbbr = jornalName
# author name, affiliationInfo
if temp.find("Article").find("AuthorList") != None:
authorList = temp.find("Article").find("AuthorList").findall(
"Author")
for i in authorList:
name_temp = []
if i.find("LastName") != None:
name_temp.append(i.find("LastName").text)
if i.find("ForeName") != None:
name_temp.append(i.find("ForeName").text)
if name_temp==[]:
authorNameList.append(["None"])
else:
authorNameList.append(name_temp)
authorAff = [[
j.find("Affiliation").text
for j in i.findall("AffiliationInfo")
] for i in authorList]
# MESH terms
if temp.find("MeshHeadingList") != None:
for i in temp.find("MeshHeadingList").findall("MeshHeading"):
# save MESH terms
MESH_allTerms.append(i.find("DescriptorName").text)
# save MESH major terms
if i.find("DescriptorName").attrib['MajorTopicYN'] == "Y":
MESH_majorTerms.append(i.find("DescriptorName").text)
# QualifierName 的属性MajorTopicYN=Y,则添加DescriptorName至 major terms
elif i.find("QualifierName") != None:
for j in i.findall("QualifierName"):
if j.attrib['MajorTopicYN'] == "Y":
MESH_majorTerms.append(
i.find("DescriptorName").text)
break
# grant
grantInfoList = []
if temp.find("Article").find("GrantList") != None:
grantList = temp.find("Article").find("GrantList")
count2 = 0
for i in grantList.findall("Grant"):
count2 += 1
# print(count2, "\n")
# GrantID, Agency, Country
GrantID = ""
Agency = ""
Country = ""
if i.find("GrantID") != None:
GrantID = i.find("GrantID").text
if i.find("Agency") != None:
Agency = i.find("Agency").text
if i.find("Country") != None:
Country = i.find("Country").text
grantInfoList.append([GrantID, Agency, Country])
# cites
# 如果CommentsCorrections,RefType=RetractionOf,代表被撤稿,去除这篇文章
if temp.find("CommentsCorrectionsList") != None:
commentList = temp.find("CommentsCorrectionsList").findall(
"CommentsCorrections")
for i in commentList:
# Cites lists items in the bibliography or list of references at the end of an article.
if list(i.attrib.values())[0] == "RetractionOf":
RetractionOf = 1
break
if list(i.attrib.values())[0] != "Cites":
continue
citedList.append([
i.find("RefSource").text.split(".")[0],
i.find("PMID").text
])
# citedList=[[i.find("RefSource").text.split(".")[0],i.find("PMID").text] for i in commentList]
# 被撤稿,跳过
if RetractionOf == 1:
continue
else:
count += 1
print(count, "\n")
pmid_arr.append(pmid)
pubData_arr.append(pubData)
articleTitle_arr.append(articleTitle)
articleAbstract_arr.append(articleAbstract)
jornalName_arr.append(jornalName)
jornalNameAbbr_arr.append(jornalNameAbbr)
authorNameList_arr.append(authorNameList)
authorAff_arr.append(authorAff)
MESH_allTerms_arr.append(MESH_allTerms)
MESH_majorTerms_arr.append(MESH_majorTerms)
grantInfoList_arr.append(grantInfoList)
citedList_arr.append(citedList)
# 添加期刊的2018影响因子
if2018_jornalName_upper=[i.upper() for i in if2018[:,0]]
for i in jornalName_arr:
flag=0
for j in np.arange(len(if2018_jornalName_upper)):
if if2018_jornalName_upper[j] == i.upper():
if2018_arr.append(if2018[j,1])
flag=1
break
elif if2018_jornalName_upper[j] in i.upper():
if2018_arr.append(if2018[j,1])
flag=1
break
if flag==0:
if2018_arr.append("None")
# 识别一作、通讯作者和所有作者的国别
firstAuthorCountryInstitute = FirstAuthorCountry(authorAff_arr,
country_name)
contectAuthorCountryInstitute = ContectAuthorCountry(
authorAff_arr, country_name)
eachAuthorCountry = EachAuthorCountry(authorAff_arr, country_name)
#将所有作者的国别拼接成字符串
eachAuthorCountry=[",".join(i) for i in eachAuthorCountry]
return ([
pmid_arr, jornalName_arr, pubData_arr, jornalNameAbbr_arr,
articleTitle_arr, articleAbstract_arr, authorNameList_arr,
authorAff_arr, MESH_allTerms_arr, MESH_majorTerms_arr,
grantInfoList_arr, citedList_arr, firstAuthorCountryInstitute[0],
firstAuthorCountryInstitute[1], contectAuthorCountryInstitute[0],
contectAuthorCountryInstitute[1], eachAuthorCountry,if2018_arr
]) 解析好文章结构以后,下面主要是数据的批量导入
二、批量导入mysql数据库
由于下载的文章篇数较大,所以采用批量导入mysql数据库的方法,在python3中,我主要采用了pymysql工具包,需要在本地配置好mysql数据库的环境以及建立表的字段信息,建立的表字段信息如下:
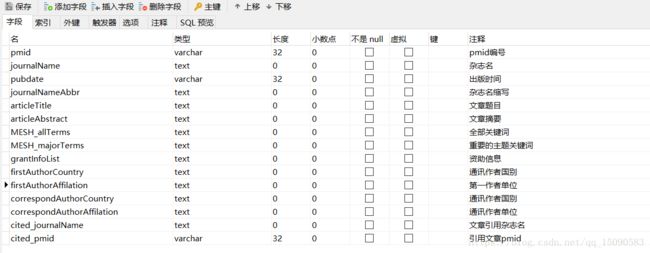
批量导入代码如下:
def db_insert():
#创建连接
conn = pymysql.Connect(
host = 'localhost',
port = 3306,
user = 'root',
passwd = '4910203',
db = 'ra_pubmed',
charset = 'utf8mb4',
cursorclass=pymysql.cursors.DictCursor
)
#获取cursor
cur = conn.cursor()
sql = 'insert into paper_info(pmid,date,journalname,journalabbr,title,abstract) values(%s,%s,%s,%s,%s,%s)'#本地数据库中的部分字段名和表名
#sql = 'insert into test(pmid,date) values(%s,%s)'
path = "E:/RA_Investigate/pubmed_RA.xml"
article_info = read_xml(path)
ls=[(article_info[0][i],article_info[1][i],article_info[2][i],article_info[3][i],article_info[4][i],article_info[5][i]) for i in np.arange(len(article_info[0]))]#这里传入的字段和上面解析的存在不一致,因为开始我只是测试。
#ls= [(article_info[0][i],article_info[1][i]) for i in np.arange(len(article_info[0])) ]
cur.executemany(sql,ls)
#提交更新
conn.commit()
#关闭连接对象
cur.close()