KEGG功能注释工具 KofamKOALA 安装与使用
KEGG数据库,即京都基因和基因组百科全书(Kyoto Encyclopedia of Genes and Genomes),是系统分析基因功能、基因组信息的数据库。
KofamKOALA是一个方便的KEGG功能注释工具,由创建KEGG的京都大学化研所生物信息中心学者在2019年11月发表于Bioinformatics。
以隐马尔科夫模型(HMM)创建的KOfam来进行蛋白序列同源搜索,其准确性可与性能最佳的工具相媲美, 有网页和Linux两个版本,本文重点介绍Linux版的安装与使用。
网页版
网址 https://www.genome.jp/tools/kofamkoala/

avatar
网页填入蛋白序列信息,设值E值和留下邮箱点击Compute,只需要等待邮箱回复
Linux版
Linux版本的KofamKOALA 需要下载 KOfam(数据库)和 KofamScan(软件),软件依赖Ruby,HMMER和GNU Parallel(事先没有安装可以看以下教程)
安装
我们以kofamscan安装在主目录$HOME(或者叫~)下为例介绍:
step1
下载和解压 KOfam 和 KofamScan
mkdir -p ~/kofamscan/dbcd ~/kofamscan/dbwget ftp://ftp.genome.jp/pub/db/kofam/ko_list.gz wget ftp://ftp.genome.jp/pub/db/kofam/profiles.tar.gz gunzip ko_list.gz tar xvzf profiles.tar.gz mkdir -p ~/kofamscan/bincd ~/kofamscan/binwget ftp://ftp.genome.jp/pub/tools/kofamscan/kofamscan-1.2.0.tar.gz # 注意kofamscan版本tar xvzf kofamscan-1.2.0.tar.gz
step2
下载 Ruby HMMER GNU Parallel
cd ~/kofamscan mkdir ruby hmmer parallel src cd src# Ruby版本应不小于2.4,这里演示的是2.7版;HMMER应大于3.1,这里是3.3;Parallel为最新版wget https://cache.ruby-lang.org/pub/ruby/2.7/ruby-2.7.0.tar.gzwget http://eddylab.org/software/hmmer/hmmer-3.3.tar.gzwget ftp://ftp.gnu.org/gnu/parallel/parallel-latest.tar.bz2
安装 Ruby
cd ~/kofamscan/src tar xvzf ruby-2.7.0.tar.gz cd ruby-2.7.0./configure --prefix=$HOME/kofamscan/ruby make make install
安装 HMMER
cd ~/kofamscan/src tar xvzf hmmer-3.3.tar.gz cd hmmer-3.3./configure --prefix=$HOME/kofamscan/hmmer make make install
安装 GNU Parallel
cd ~/kofamscan/src tar xvjf parallel-latest.tar.bz2 cd parallel-20190322 # 这里根据版本而定./configure --prefix=$HOME/kofamscan/parallel make make install
将Ruby路径加入环境变量(之后执行如果报错可能是ruby的问题,推荐ruby按照本文方法安装)
export PATH=$HOME/kofamscan/ruby/bin:$PATH
step3
复制config-template.yml文件并重命名为config.yml
cd ~/kofamscan/bin/ cp config-template.yml config.yml
cat config.yml文件,内容如下:
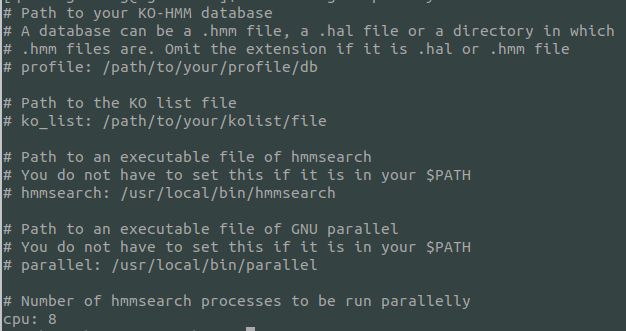
我们需要在config.yml添加键值,以便ruby识别读取.
可以vim编辑加入以下内容到config.yml,注意绝对路径.
profile: /path/to/home/kofamscan/db/profiles ko_list: /path/to/home/kofamscan/db/ko_list hmmsearch: /path/to/home/kofamscan/hmmer/bin/hmmsearch parallel: /path/to/home/kofamscan/parallel/bin/parallel
如:
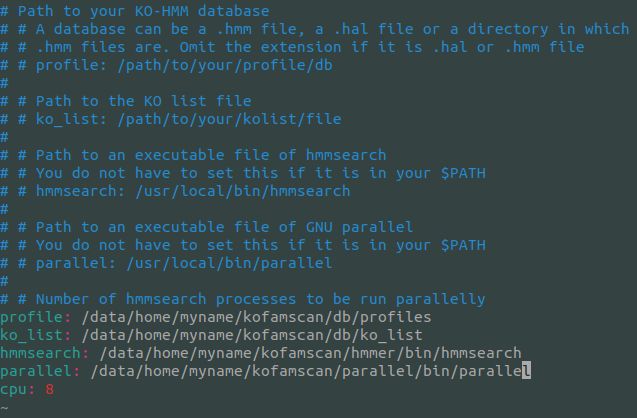
若hummsearch和parallel可安装在其他地方改为相关路径
使用
现在可以使用了,我们准备蛋白序列fasta文件(注意必须是蛋白序列,不支持核酸序列)
./exec_annotation -o result.txt query.fasta
如我在~/kofamscan/test/文件夹下有samples.fasta文件,定义输出文件为res.txt
cd ~/kofamscan/bin/kofamscan-1.2.0 # 路径中有exec_annotation文件./exec_annotation -o ~/kofamscan/test/res.txt~/kofamscan/test/samples.fasta
运行完毕后的输出文件:
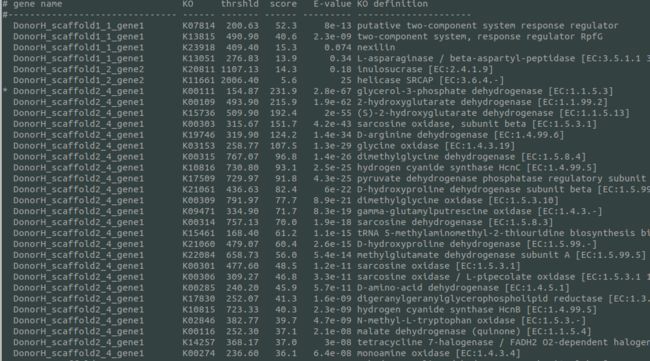
若报错可能是ruby的路径不在首选环境变量,可执行:
export PATH=$HOME/kofamscan/ruby/bin:$PATH
./exec_annotation -h查看全部参数:
## Options
- `-o FILE`
- The result are output to `FILE`. It defaults to `stdout`.
- `-p`, `--profile=PROFILE`
- Use `PROFILE` as a profile database. See [Profiles](#profiles)
- `-k`, `--ko-list=FILE`
- Use `FILE` as a KO list.
- `--cpu=N`
- Set the number of `hmmsearch` processes started simultaneously to `N`. It defaults to 1 unless it is set in `config.yml`.
- `-c FILE`
- Use `FILE` as a config file instead of `config.yml` in the same directory as `exec_annotation`.
- `--tmp-dir=DIR`
- Use `DIR` as a temporary directory where hmmsearch results are. It will be created if not exist. It defaults to `./tmp`.
- `-E`, `--e-value=VALUE`
- Require E-value to be smaller than or equal to `VALUE`. If not, an asterisk will not be added in `detail` format or the hit will not be reported in other formats.
- `-T`, `--threshold-scale=VALUE`
- The score thresholds are multiplied by `VALUE`. For example, with `-T2` option, the thresholds become twice as strict.
- `-f`, `--format=FORMAT`
- Set the format of the output to `FORMAT`. Three formats below are available.
- `detail`
- Default format. Gene name, assigned K number, threshold of the KO, hmmsearch score and E-value, and the definition of KO are shown. In addition, an asterisk '*' is added to the head of the line if the score is higher than the threshold.
- `mapper`
- Format which can be used for [KEGG Mapper](https://www.genome.jp/kegg/mapper.html) input. It includes a gene name and an assigned K number separated by a tab. Here, an assigned K number represents a hit with score above the predefined threshold. Note that for some KOs, predefined score thresholds are not available when they are represented by a very few number of sequences in KEGG GENES.
- `mapper-oneline`
- Similar to `mapper`, but when more than one KO are assigned to a gene, all assigned KO are shown in one line separated by tabs.
- `--[no-]report-unannotated`
- With `--report-unannotated` option, gene names are shown even when no KO is assigned (default when `--format=mapper(-oneline)`). With `--no-report-unannotated` such genes are not shown at all (default when `--format=detail`).
- `--create-alignment`
- `hmmsearch`'s normal outputs per profile are stored in the temporary directory. In addition, domain information and alignments in the outputs will be rearranged per query.
- Not compatible with `--reannotation`
- `-r`, `--reannotation`
- Skip `hmmsearch` and assume that `hmmsearch` outputs are already in the temporary directory. This will help you to make an output in a different format or redo annotation changing thresholds.
- Not compatible with `--create-alignment`
- `-h`, `--help`
- Show brief help message.
参考 :
https://www.genome.jp/tools/kofamkoala/
https://academic.oup.com/bioinformatics/advance-article/doi/10.1093/bioinformatics/btz859/5631907
猜你喜欢
10000+:菌群分析 宝宝与猫狗 梅毒狂想曲 提DNA发Nature Cell专刊 肠道指挥大脑
系列教程:微生物组入门 Biostar 微生物组 宏基因组
专业技能:学术图表 高分文章 生信宝典 不可或缺的人
一文读懂:宏基因组 寄生虫益处 进化树
必备技能:提问 搜索 Endnote
文献阅读 热心肠 SemanticScholar Geenmedical
扩增子分析:图表解读 分析流程 统计绘图
16S功能预测 PICRUSt FAPROTAX Bugbase Tax4Fun
在线工具:16S预测培养基 生信绘图
科研经验:云笔记 云协作 公众号
编程模板: Shell R Perl
生物科普: 肠道细菌 人体上的生命 生命大跃进 细胞暗战 人体奥秘
写在后面
为鼓励读者交流、快速解决科研困难,我们建立了“宏基因组”专业讨论群,目前己有国内外5000+ 一线科研人员加入。参与讨论,获得专业解答,欢迎分享此文至朋友圈,并扫码加主编好友带你入群,务必备注“姓名-单位-研究方向-职称/年级”。PI请明示身份,另有海内外微生物相关PI群供大佬合作交流。技术问题寻求帮助,首先阅读《如何优雅的提问》学习解决问题思路,仍未解决群内讨论,问题不私聊,帮助同行。
![]()
学习16S扩增子、宏基因组科研思路和分析实战,关注“宏基因组”
![]() 点击阅读原文
点击阅读原文