ggalluvial:冲击图展示组间变化、时间序列和复杂多属性alluvial diagram
感谢“宏基因组0”群友李海敏、沈伟推荐此包绘制堆叠柱状图各成分连线:突出展示组间物种丰度变化。
冲击图(alluvial diagram)是流程图(flow diagram)的一种,最初开发用于代表网络结构的时间变化。
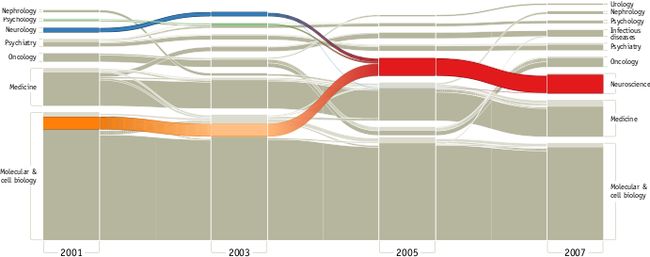
实例1. neuroscience coalesced from other related disciplines to form its own field. From PLoS ONE 5(1): e8694 (2010)
![]()
实例2. Sciences封面哈扎人肠道菌群 图1中的C/D就使用了3个冲击图。详见3分和30分文章差距在哪里?
ggalluvial是一个基于ggplot2的扩展包,专门用于快速绘制冲击图(alluvial diagram),有些人也叫它桑基图(Sankey diagram),但两者略有区别,将来我们会介绍riverplot包绘制桑基图。
软件源代码位于Github: https://github.com/corybrunson/ggalluvial
CRNA官方演示教程: https://cran.r-project.org/web/packages/ggalluvial/vignettes/ggalluvial.html
安装
以下三种方装方式,三选1:
# 国内用户推荐清华镜像站
site="https://mirrors.tuna.tsinghua.edu.cn/CRAN"
# 安装稳定版(推荐)
install.packages("ggalluvial", repo=site)
# 安装开发版(连github不稳定有时间下载失败,多试几次可以成功)
devtools::install_github("corybrunson/ggalluvial", build_vignettes = TRUE)
# 安装新功能最优版
devtools::install_github("corybrunson/ggalluvial", ref = "optimization")显示帮助文档
使用vignette查看演示教程
# 查看教程
vignette(topic = "ggalluvial", package = "ggalluvial")接下来我们的演示均基于此官方演示教程,我的主要贡献是翻译与代码注释。
基于ggplot2的冲击图
原作者:Jason Cory Brunson, 更新日期:2018-02-11
1. 最简单的示例
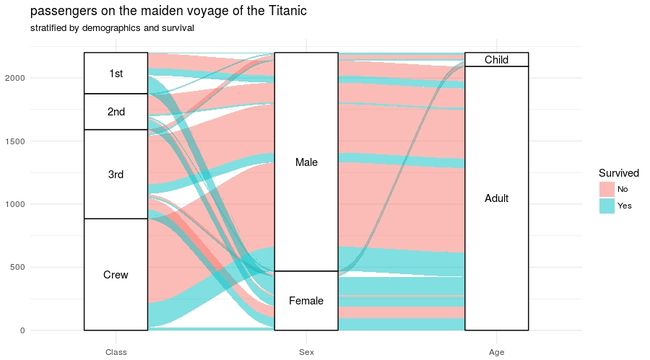
基于泰坦尼克事件人员统计绘制性别与舱位和年龄的关系。
# 加载包
library(ggalluvial)
# 转换内部数据为数据框,宽表格模式
titanic_wide <- data.frame(Titanic)
# 显示数据格式
head(titanic_wide)
#> Class Sex Age Survived Freq
#> 1 1st Male Child No 0
#> 2 2nd Male Child No 0
#> 3 3rd Male Child No 35
#> 4 Crew Male Child No 0
#> 5 1st Female Child No 0
#> 6 2nd Female Child No 0
# 绘制性别与舱位和年龄的关系
ggplot(data = titanic_wide,
aes(axis1 = Class, axis2 = Sex, axis3 = Age,
weight = Freq)) +
scale_x_discrete(limits = c("Class", "Sex", "Age"), expand = c(.1, .05)) +
geom_alluvium(aes(fill = Survived)) +
geom_stratum() + geom_text(stat = "stratum", label.strata = TRUE) +
theme_minimal() +
ggtitle("passengers on the maiden voyage of the Titanic",
"stratified by demographics and survival")具体参考说明:data设置数据源,axis设置显示的柱,weight为数值,geom_alluvium为冲击图组间面积连接并按生存率比填充分组,geom_stratum()每种有柱状图,geom_text()显示柱状图中标签,theme_minimal()主题样式的一种,ggtitle()设置图标题
图1. 展示性别与舱位和年龄的关系及存活率比例
我们发现上图居然画的是宽表格模式下的数据,而通常ggplot2处理都是长表格模式,如何转换呢?
to_loades转换为长表格
# 长表格模式,to_loades多组组合,会生成alluvium和stratum列。主分组位于命名的key列中
titanic_long <- to_lodes(data.frame(Titanic),
key = "Demographic",
axes = 1:3)
head(titanic_long)
ggplot(data = titanic_long,
aes(x = Demographic, stratum = stratum, alluvium = alluvium,
weight = Freq, label = stratum)) +
geom_alluvium(aes(fill = Survived)) +
geom_stratum() + geom_text(stat = "stratum") +
theme_minimal() +
ggtitle("passengers on the maiden voyage of the Titanic",
"stratified by demographics and survival")产生和上图一样的图,只是数据源格式不同。
2. 输入数据格式
定义一种Alluvial宽表格
# 显示数据格式
head(as.data.frame(UCBAdmissions), n = 12)
## Admit Gender Dept Freq
## 1 Admitted Male A 512
## 2 Rejected Male A 313
## 3 Admitted Female A 89
## 4 Rejected Female A 19
## 5 Admitted Male B 353
## 6 Rejected Male B 207
## 7 Admitted Female B 17
## 8 Rejected Female B 8
## 9 Admitted Male C 120
## 10 Rejected Male C 205
## 11 Admitted Female C 202
## 12 Rejected Female C 391
# 判断数据格式
is_alluvial(as.data.frame(UCBAdmissions), logical = FALSE, silent = TRUE)
## [1] "alluvia"查看性别与专业间关系,并按录取情况分组
ggplot(as.data.frame(UCBAdmissions),
aes(weight = Freq, axis1 = Gender, axis2 = Dept)) +
geom_alluvium(aes(fill = Admit), width = 1/12) +
geom_stratum(width = 1/12, fill = "black", color = "grey") +
geom_label(stat = "stratum", label.strata = TRUE) +
scale_x_continuous(breaks = 1:2, labels = c("Gender", "Dept")) +
scale_fill_brewer(type = "qual", palette = "Set1") +
ggtitle("UC Berkeley admissions and rejections, by sex and department")
```

### 3. 三类型间关系,按重点着色
Titanic按生存,性别,舱位分类查看关系,并按舱位填充色
ggplot(as.data.frame(Titanic),
aes(weight = Freq,
axis1 = Survived, axis2 = Sex, axis3 = Class)) +
geom_alluvium(aes(fill = Class),
width = 0, knot.pos = 0, reverse = FALSE) +
guides(fill = FALSE) +
geom_stratum(width = 1/8, reverse = FALSE) +
geom_text(stat = “stratum”, label.strata = TRUE, reverse = FALSE) +
scale_x_continuous(breaks = 1:3, labels = c(“Survived”, “Sex”, “Class”)) +
coord_flip() +
ggtitle(“Titanic survival by class and sex”)
“`

4. 长表格数据
# to_lodes转换为长表格
UCB_lodes <- to_lodes(as.data.frame(UCBAdmissions), axes = 1:3)
head(UCB_lodes, n = 12)
## Freq alluvium x stratum
## 1 512 1 Admit Admitted
## 2 313 2 Admit Rejected
## 3 89 3 Admit Admitted
## 4 19 4 Admit Rejected
## 5 353 5 Admit Admitted
## 6 207 6 Admit Rejected
## 7 17 7 Admit Admitted
## 8 8 8 Admit Rejected
## 9 120 9 Admit Admitted
## 10 205 10 Admit Rejected
## 11 202 11 Admit Admitted
## 12 391 12 Admit Rejected
# 判断是否符合格式要求
is_alluvial(UCB_lodes, logical = FALSE, silent = TRUE)
## [1] "alluvia"
主要列说明:
- x, 主要的分类,即X轴上每个柱
- stratum, 主要分类中的分组
- alluvium, 连接图的索引
5. 绘制非等高冲击图
以各国难民数据为例,观察多国难民数量随时间变化
data(Refugees, package = "alluvial")
country_regions <- c(
Afghanistan = "Middle East",
Burundi = "Central Africa",
`Congo DRC` = "Central Africa",
Iraq = "Middle East",
Myanmar = "Southeast Asia",
Palestine = "Middle East",
Somalia = "Horn of Africa",
Sudan = "Central Africa",
Syria = "Middle East",
Vietnam = "Southeast Asia"
)
Refugees$region <- country_regions[Refugees$country]
ggplot(data = Refugees,
aes(x = year, weight = refugees, alluvium = country)) +
geom_alluvium(aes(fill = country, colour = country),
alpha = .75, decreasing = FALSE) +
scale_x_continuous(breaks = seq(2003, 2013, 2)) +
theme(axis.text.x = element_text(angle = -30, hjust = 0)) +
scale_fill_brewer(type = "qual", palette = "Set3") +
scale_color_brewer(type = "qual", palette = "Set3") +
facet_wrap(~ region, scales = "fixed") +
ggtitle("refugee volume by country and region of origin")
```

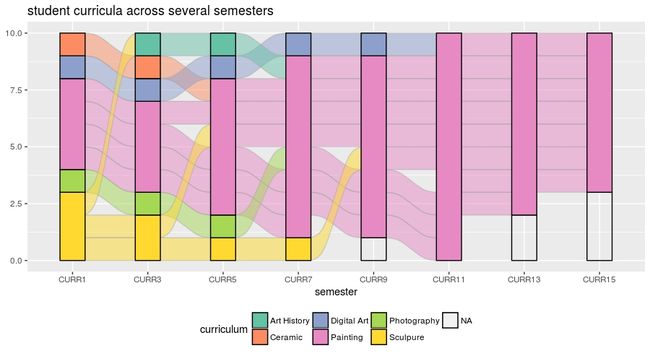
### 6. 等高非等量关系
不同学期学生学习科目的变化
data(majors)
majors curriculum<−as.factor(majors c u r r i c u l u m < − a s . f a c t o r ( m a j o r s curriculum)
ggplot(majors,
aes(x = semester, stratum = curriculum, alluvium = student,
fill = curriculum, label = curriculum)) +
scale_fill_brewer(type = “qual”, palette = “Set2”) +
geom_flow(stat = “alluvium”, lode.guidance = “rightleft”,
color = “darkgray”) +
geom_stratum() +
theme(legend.position = “bottom”) +
ggtitle(“student curricula across several semesters”)
“`
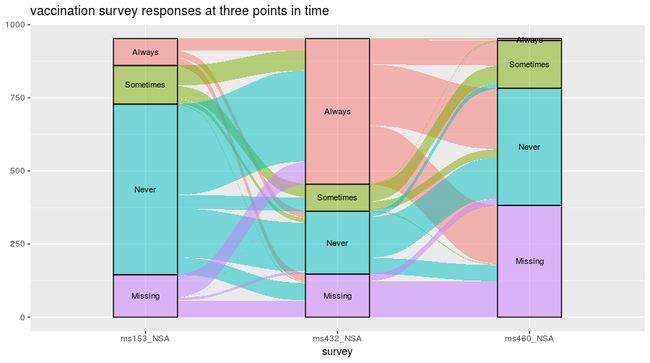
7. 工作状态时间变化图
data(vaccinations)
levels(vaccinations$response) <- rev(levels(vaccinations$response))
ggplot(vaccinations,
aes(x = survey, stratum = response, alluvium = subject,
weight = freq,
fill = response, label = response)) +
geom_flow() +
geom_stratum(alpha = .5) +
geom_text(stat = "stratum", size = 3) +
theme(legend.position = "none") +
ggtitle("vaccination survey responses at three points in time")8. 分类学门水平相对丰度实战
# 实战1. 组间丰度变化
# 编写测试数据
df=data.frame(
Phylum=c("Ruminococcaceae","Bacteroidaceae","Eubacteriaceae","Lachnospiraceae","Porphyromonadaceae"),
GroupA=c(37.7397,31.34317,222.08827,5.08956,3.7393),
GroupB=c(113.2191,94.02951,66.26481,15.26868,11.2179),
GroupC=c(123.2191,94.02951,46.26481,35.26868,1.2179),
GroupD=c(37.7397,31.34317,222.08827,5.08956,3.7393)
)
# 数据转换长表格
library(reshape2)
melt_df = melt(df)
# 绘制分组对应的分类学,有点像circos
ggplot(data = melt_df,
aes(axis1 = Phylum, axis2 = variable,
weight = value)) +
scale_x_discrete(limits = c("Phylum", "variable"), expand = c(.1, .05)) +
geom_alluvium(aes(fill = Phylum)) +
geom_stratum() + geom_text(stat = "stratum", label.strata = TRUE) +
theme_minimal() +
ggtitle("Phlyum abundance in each group")绘制分组对应的分类学,有点像circos
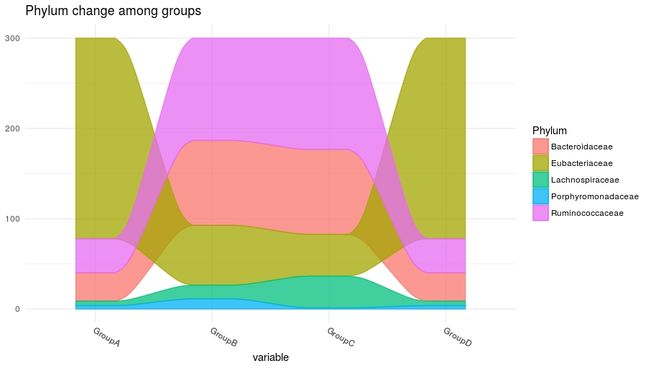
# 组间各丰度变化
ggplot(data = melt_df,
aes(x = variable, weight = value, alluvium = Phylum)) +
geom_alluvium(aes(fill = Phylum, colour = Phylum, colour = Phylum),
alpha = .75, decreasing = FALSE) +
theme_minimal() +
theme(axis.text.x = element_text(angle = -30, hjust = 0)) +
ggtitle("Phylum change among groups")组间各丰度变化,如果组为时间效果更好
Reference
# 如何引用
citation("ggalluvial")Jason Cory Brunson (2017). ggalluvial: Alluvial Diagrams in ‘ggplot2’. R package version 0.5.0.
https://CRAN.R-project.org/package=ggalluvial
https://en.wikipedia.org/wiki/Alluvial_diagram
ggalluvial包源码:http://corybrunson.github.io/ggalluvial/index.html
官方示例 Alluvial Diagrams in ggplot2 https://cran.r-project.org/web/packages/ggalluvial/vignettes/ggalluvial.html
猜你喜欢
- 热文:1高分文章 2不可或缺的人 3图表规范
- 一文读懂:1微生物组 2寄生虫益处 3进化树
- 必备技能:1提问 2搜索 3Endnote
- 文献阅读 1热心肠 2SemanticScholar 3geenmedical
- 扩增子分析:1图表解读 2分析流程 3统计绘图 4功能预测
- 科研经验:1云笔记 2云协作 3公众号
- 系列教程:1Biostar 2微生物组 3宏基因组
- 生物科普 1肠道细菌 2人体上的生命 3生命大跃进 4细胞的暗战 5人体奥秘
写在后面
为鼓励读者交流、快速解决科研困难,我们建立了“宏基因组”专业讨论群,目前己有国内外1200+ 一线科研人员加入。参与讨论,获得专业解答,欢迎分享此文至朋友圈,并扫码加主编好友带你入群,务必备注“姓名-单位-研究方向-职称/年级”。技术问题寻求帮助,首先阅读《如何优雅的提问》学习解决问题思路,仍末解决群内讨论,问题不私聊,帮助同行。
![]()
学习扩增子、宏基因组科研思路和分析实战,关注“宏基因组”
![]()
点击阅读原文,跳转最新文章目录阅读
https://mp.weixin.qq.com/s/5jQspEvH5_4Xmart22gjMA