cs231n assignment1 KNN分类器
# Run some setup code for this notebook.
import random
import numpy as np
from cs231n.data_utils import load_CIFAR10
import matplotlib.pyplot as plt
#from __future__ import print_function
# This is a bit of magic to make matplotlib figures appear inline in the notebook
# rather than in a new window.
%matplotlib inline
plt.rcParams['figure.figsize'] = (10.0, 8.0) # set default size of plots
plt.rcParams['image.interpolation'] = 'nearest'
plt.rcParams['image.cmap'] = 'gray'
# Some more magic so that the notebook will reload external python modules;
# see http://stackoverflow.com/questions/1907993/autoreload-of-modules-in-ipython
%load_ext autoreload
%autoreload 2
第一个问题:

老是跳出这个不能执行
解决方法就是等这里加载完 …

第二个问题:

不知道为什么会出现这个 明明放在正确的文件下 如果大家有同样的问题 跪求指点!!
只能自己添加路径了
import os
os.chdir("E:/CatchBeliF/study/homework/assignment1/")
print(os.getcwd())
# Load the raw CIFAR-10 data.
cifar10_dir = 'cs231n/datasets/cifar-10-batches-py'
X_train, y_train, X_test, y_test = load_CIFAR10(cifar10_dir)
# As a sanity check, we print out the size of the training and test data.
print('Training data shape: ', X_train.shape)
print('Training labels shape: ', y_train.shape)
print('Test data shape: ', X_test.shape)
print('Test labels shape: ', y_test.shape)
Training data shape: (50000, 32, 32, 3)
Training labels shape: (50000,)
Test data shape: (10000, 32, 32, 3)
Test labels shape: (10000,)
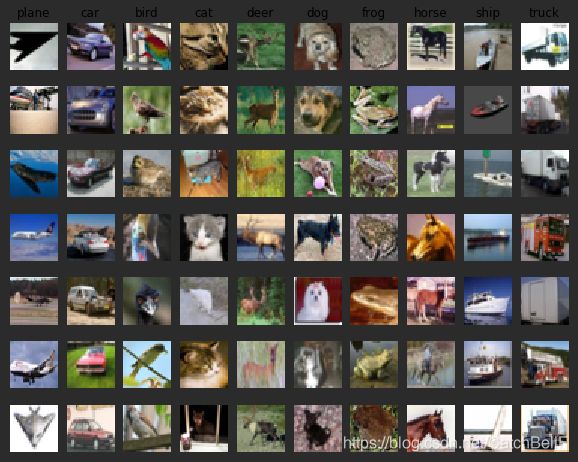
# Visualize some examples from the dataset.
# We show a few examples of training images from each class.
classes = ['plane', 'car', 'bird', 'cat', 'deer', 'dog', 'frog', 'horse', 'ship', 'truck']
num_classes = len(classes)
samples_per_class = 7 #每个类别采样个数
for y, cls in enumerate(classes):
idxs = np.flatnonzero(y_train == y) #找出标签中y类的位置
idxs = np.random.choice(idxs, samples_per_class, replace=False)#从中选出7个样本
for i, idx in enumerate(idxs):#对所选的样本的位置和样本所对应的图片在训练集中的位置进行循环
plt_idx = i * num_classes + y + 1#在子图中所占位置的计算
plt.subplot(samples_per_class, num_classes, plt_idx)#说明要画的子图的编号
plt.imshow(X_train[idx].astype('uint8'))
plt.axis('off')
if i == 0:
plt.title(cls)
plt.show()
补充
其中enumerate() 函数用于将一个可遍历的数据对象(如列表、元组或字符串)组合为一个索引序列,同时列出数据和数据下标

继续执行代码
# Subsample the data for more efficient code execution in this exercise
num_training = 5000
mask = list(range(num_training))
X_train = X_train[mask]
y_train = y_train[mask]
num_test = 500
mask = list(range(num_test))
X_test = X_test[mask]
y_test = y_test[mask]
# Reshape the image data into rows
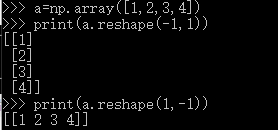
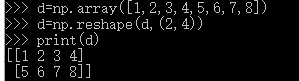
X_train = np.reshape(X_train, (X_train.shape[0], -1))
X_test = np.reshape(X_test, (X_test.shape[0], -1))
print(X_train.shape, X_test.shape)
(5000, 3072) (500, 3072)
补充
其中reshape函数()
一个参数为-1时,那么reshape函数会根据另一个参数的维度计算出数组的另外一个shape属性值。

继续执行代码
from cs231n.classifiers import KNearestNeighbor
# Create a kNN classifier instance.
# Remember that training a kNN classifier is a noop:
# the Classifier simply remembers the data and does no further processing
classifier = KNearestNeighbor()
classifier.train(X_train, y_train)
补充
knn分类器的主要步骤:
1.训练。
读取训练数据并存储。
2.测试。
对于每一张测试图像,kNN把它与训练集中的每一张图像计算距离,找出距离最近的k张图像.这k张图像里,占多数的标签类别,就是测试图像的类别。.
继续执行代码
# Open cs231n/classifiers/k_nearest_neighbor.py and implement
# compute_distances_two_loops.
# Test your implementation:
dists = classifier.compute_distances_two_loops(X_test)
print(dists.shape)
(500, 5000)
# We can visualize the distance matrix: each row is a single test example and
# its distances to training examples
plt.imshow(dists, interpolation='none')
plt.show()
一开始显示全黑后来莫名就好了 有同样情况的朋友请告知啊!


深色表示距离小,浅色表示距离大
某些行颜色偏浅,表示测试样本与训练集中的所有样本差异较大,该测试样本可能明显过亮或过暗或者有色差。
某些列颜色偏浅,所有测试样本与该列表示的训练样本距离都较大,该训练样本可能明显过亮或过暗或者有色差。
继续执行代码
# Now implement the function predict_labels and run the code below:
# We use k = 1 (which is Nearest Neighbor).
y_test_pred = classifier.predict_labels(dists, k=1)
# Compute and print the fraction of correctly predicted examples
num_correct = np.sum(y_test_pred == y_test)
accuracy = float(num_correct) / num_test
print('Got %d / %d correct => accuracy: %f' % (num_correct, num_test, accuracy)
Got 137 / 500 correct => accuracy: 0.274000
y_test_pred = classifier.predict_labels(dists, k=5)
num_correct = np.sum(y_test_pred == y_test)
accuracy = float(num_correct) / num_test
print('Got %d / %d correct => accuracy: %f' % (num_correct, num_test, accuracy))
Got 139 / 500 correct => accuracy: 0.278000
# Now lets speed up distance matrix computation by using partial vectorization
# with one loop. Implement the function compute_distances_one_loop and run the
# code below:
dists_one = classifier.compute_distances_one_loop(X_test)
# To ensure that our vectorized implementation is correct, we make sure that it
# agrees with the naive implementation. There are many ways to decide whether
# two matrices are similar; one of the simplest is the Frobenius norm. In case
# you haven't seen it before, the Frobenius norm of two matrices is the square
# root of the squared sum of differences of all elements; in other words, reshape
# the matrices into vectors and compute the Euclidean distance between them.
difference = np.linalg.norm(dists - dists_one, ord='fro')
print('Difference was: %f' % (difference, ))
if difference < 0.001:
print('Good! The distance matrices are the same')
else:
print('Uh-oh! The distance matrices are different')
Difference was: 0.000000
Good! The distance matrices are the same
# Now implement the fully vectorized version inside compute_distances_no_loops
# and run the code
dists_two = classifier.compute_distances_no_loops(X_test)
# check that the distance matrix agrees with the one we computed before:
difference = np.linalg.norm(dists - dists_two, ord='fro')
print('Difference was: %f' % (difference, ))
if difference < 0.001:
print('Good! The distance matrices are the same')
else:
print('Uh-oh! The distance matrices are different')
Difference was: 0.000000
Good! The distance matrices are the same
# Let's compare how fast the implementations are
def time_function(f, *args):
"""
Call a function f with args and return the time (in seconds) that it took to execute.
"""
import time
tic = time.time()
f(*args)
toc = time.time()
return toc - tic
two_loop_time = time_function(classifier.compute_distances_two_loops, X_test)
print('Two loop version took %f seconds' % two_loop_time)
one_loop_time = time_function(classifier.compute_distances_one_loop, X_test)
print('One loop version took %f seconds' % one_loop_time)
no_loop_time = time_function(classifier.compute_distances_no_loops, X_test)
print('No loop version took %f seconds' % no_loop_time)
# you should see significantly faster performance with the fully vectorized implementati
Two loop version took 87.875775 seconds
One loop version took 137.346529 seconds
No loop version took 0.785548 seconds
num_folds = 5
k_choices = [1, 3, 5, 8, 10, 12, 15, 20, 50, 100]
X_train_folds = []
y_train_folds = []
################################################################################
# TODO: #
# Split up the training data into folds. After splitting, X_train_folds and #
# y_train_folds should each be lists of length num_folds, where #
# y_train_folds[i] is the label vector for the points in X_train_folds[i]. #
# Hint: Look up the numpy array_split function. #
################################################################################
X_train_folds=np.array_split(X_train,num_folds)
y_train_folds=np.array_split(y_train,num_folds)
################################################################################
# END OF YOUR CODE #
################################################################################
# A dictionary holding the accuracies for different values of k that we find
# when running cross-validation. After running cross-validation,
# k_to_accuracies[k] should be a list of length num_folds giving the different
# accuracy values that we found when using that value of k.
k_to_accuracies = {}
################################################################################
# TODO: #
# Perform k-fold cross validation to find the best value of k. For each #
# possible value of k, run the k-nearest-neighbor algorithm num_folds times, #
# where in each case you use all but one of the folds as training data and the #
# last fold as a validation set. Store the accuracies for all fold and all #
# values of k in the k_to_accuracies dictionary. #
################################################################################
classifier = KNearestNeighbor()
for k in k_choices:
accuracies = np.zeros(num_folds)
for fold in range(num_folds):
temp_X = X_train_folds[:]
temp_y = y_train_folds[:]
X_validate_fold = temp_X.pop(fold)
y_validate_fold = temp_y.pop(fold)
temp_X = np.array([y for x in temp_X for y in x])
temp_y = np.array([y for x in temp_y for y in x])
classifier.train(temp_X, temp_y)
y_test_pred = classifier.predict(X_validate_fold, k=k)
num_correct = np.sum(y_test_pred == y_validate_fold)
accuracy = float(num_correct) / num_test
accuracies[fold] =accuracy
k_to_accuracies[k] = accuracies
################################################################################
# END OF YOUR CODE #
################################################################################
# Print out the computed accuracies
for k in sorted(k_to_accuracies):
for accuracy in k_to_accuracies[k]:
print('k = %d, accuracy = %f' % (k, accuracy))
k = 1, accuracy = 0.526000
k = 1, accuracy = 0.514000
k = 1, accuracy = 0.528000
k = 1, accuracy = 0.556000
k = 1, accuracy = 0.532000
k = 3, accuracy = 0.478000
k = 3, accuracy = 0.498000
k = 3, accuracy = 0.480000
k = 3, accuracy = 0.532000
k = 3, accuracy = 0.508000
k = 5, accuracy = 0.496000
k = 5, accuracy = 0.532000
k = 5, accuracy = 0.560000
k = 5, accuracy = 0.584000
k = 5, accuracy = 0.560000
k = 8, accuracy = 0.524000
k = 8, accuracy = 0.564000
k = 8, accuracy = 0.546000
k = 8, accuracy = 0.580000
k = 8, accuracy = 0.546000
k = 10, accuracy = 0.530000
k = 10, accuracy = 0.592000
k = 10, accuracy = 0.552000
k = 10, accuracy = 0.568000
k = 10, accuracy = 0.560000
k = 12, accuracy = 0.520000
k = 12, accuracy = 0.590000
k = 12, accuracy = 0.558000
k = 12, accuracy = 0.566000
k = 12, accuracy = 0.560000
k = 15, accuracy = 0.504000
k = 15, accuracy = 0.578000
k = 15, accuracy = 0.556000
k = 15, accuracy = 0.564000
k = 15, accuracy = 0.548000
k = 20, accuracy = 0.540000
k = 20, accuracy = 0.558000
k = 20, accuracy = 0.558000
k = 20, accuracy = 0.564000
k = 20, accuracy = 0.570000
k = 50, accuracy = 0.542000
k = 50, accuracy = 0.576000
k = 50, accuracy = 0.556000
k = 50, accuracy = 0.538000
k = 50, accuracy = 0.532000
k = 100, accuracy = 0.512000
k = 100, accuracy = 0.540000
k = 100, accuracy = 0.526000
k = 100, accuracy = 0.512000
k = 100, accuracy = 0.526000
# plot the raw observations
for k in k_choices:
accuracies = k_to_accuracies[k]
plt.scatter([k] * len(accuracies), accuracies)
# plot the trend line with error bars that correspond to standard deviation
accuracies_mean = np.array([np.mean(v) for k,v in sorted(k_to_accuracies.items())])
accuracies_std = np.array([np.std(v) for k,v in sorted(k_to_accuracies.items())])
plt.errorbar(k_choices, accuracies_mean, yerr=accuracies_std)
plt.title('Cross-validation on k')
plt.xlabel('k')
plt.ylabel('Cross-validation accuracy')
plt.show()
# Based on the cross-validation results above, choose the best value for k,
# retrain the classifier using all the training data, and test it on the test
# data. You should be able to get above 28% accuracy on the test data.
best_k = 1
classifier = KNearestNeighbor()
classifier.train(X_train, y_train)
y_test_pred = classifier.predict(X_test, k=best_k)
# Compute and display the accuracy
num_correct = np.sum(y_test_pred == y_test)
accuracy = float(num_correct) / num_test
print('Got %d / %d correct => accuracy: %f' % (num_correct, num_test, accuracy))
Got 137 / 500 correct => accuracy: 0.274000
下面是knn分类器代码
import numpy as np
from past.builtins import xrange
class KNearestNeighbor(object):
""" a kNN classifier with L2 distance """
def __init__(self):
pass
def train(self, X, y):
"""
Train the classifier. For k-nearest neighbors this is just
memorizing the training data.
Inputs:
- X: A numpy array of shape (num_train, D) containing the training data
consisting of num_train samples each of dimension D.
- y: A numpy array of shape (N,) containing the training labels, where
y[i] is the label for X[i].
"""
self.X_train = X
self.y_train = y
def predict(self, X, k=1, num_loops=0):
"""
Predict labels for test data using this classifier.
Inputs:
- X: A numpy array of shape (num_test, D) containing test data consisting
of num_test samples each of dimension D.
- k: The number of nearest neighbors that vote for the predicted labels.
- num_loops: Determines which implementation to use to compute distances
between training points and testing points.
Returns:
- y: A numpy array of shape (num_test,) containing predicted labels for the
test data, where y[i] is the predicted label for the test point X[i].
"""
if num_loops == 0:
dists = self.compute_distances_no_loops(X)
elif num_loops == 1:
dists = self.compute_distances_one_loop(X)
elif num_loops == 2:
dists = self.compute_distances_two_loops(X)
else:
raise ValueError('Invalid value %d for num_loops' % num_loops)
return self.predict_labels(dists, k=k)
def compute_distances_two_loops(self, X):
"""
Compute the distance between each test point in X and each training point
in self.X_train using a nested loop over both the training data and the
test data.
Inputs:
- X: A numpy array of shape (num_test, D) containing test data.
Returns:
- dists: A numpy array of shape (num_test, num_train) where dists[i, j]
is the Euclidean distance between the ith test point and the jth training
point.
"""
num_test = X.shape[0]
num_train = self.X_train.shape[0]
dists = np.zeros((num_test, num_train))
for i in xrange(num_test):
for j in xrange(num_train):
#####################################################################
# TODO: #
# Compute the l2 distance between the ith test point and the jth #
# training point, and store the result in dists[i, j]. You should #
# not use a loop over dimension. #
#####################################################################
dists[i][j]=np.sqrt(np.sum(np.square(X[i]-self.X_train[j])))
#####################################################################
# END OF YOUR CODE #
#####################################################################
return dists
def compute_distances_one_loop(self, X):
"""
Compute the distance between each test point in X and each training point
in self.X_train using a single loop over the test data.
Input / Output: Same as compute_distances_two_loops
"""
num_test = X.shape[0]
num_train = self.X_train.shape[0]
dists = np.zeros((num_test, num_train))
for i in xrange(num_test):
#######################################################################
# TODO: #
# Compute the l2 distance between the ith test point and all training #
# points, and store the result in dists[i, :]. #
#######################################################################
dists[i,:]=np.sqrt(np.sum(np.square(X[i,:]-self.X_train[:]),axis=1))
#######################################################################
# END OF YOUR CODE #
#######################################################################
return dists
def compute_distances_no_loops(self, X):
"""
Compute the distance between each test point in X and each training point
in self.X_train using no explicit loops.
Input / Output: Same as compute_distances_two_loops
"""
num_test = X.shape[0]
num_train = self.X_train.shape[0]
dists = np.zeros((num_test, num_train))
#########################################################################
# TODO: #
# Compute the l2 distance between all test points and all training #
# points without using any explicit loops, and store the result in #
# dists. #
# #
# You should implement this function using only basic array operations; #
# in particular you should not use functions from scipy. #
# #
# HINT: Try to formulate the l2 distance using matrix multiplication #
# and two broadcast sums. #
#########################################################################
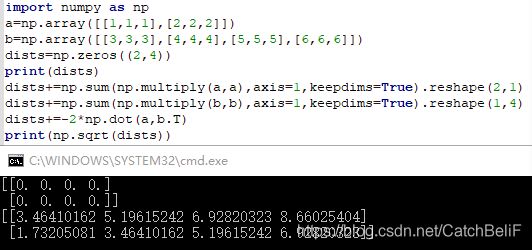
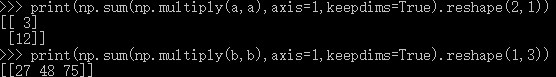
dists+=np.sum(np.multiply(X,X),axis=1,keepdims=True).reshape(num_test,1)
dists+=np.sum(np.multiply(self.X_train,self.X_train),axis=1,keepdims=True).reshape(1,num_train)
dists+=-2*np.dot(X,self.X_train.T)
dists=np.sqrt(dists)
#########################################################################
# END OF YOUR CODE #
#########################################################################
return dists
def predict_labels(self, dists, k=1):
"""
Given a matrix of distances between test points and training points,
predict a label for each test point.
Inputs:
- dists: A numpy array of shape (num_test, num_train) where dists[i, j]
gives the distance betwen the ith test point and the jth training point.
Returns:
- y: A numpy array of shape (num_test,) containing predicted labels for the
test data, where y[i] is the predicted label for the test point X[i].
"""
num_test = dists.shape[0]
y_pred = np.zeros(num_test)
for i in xrange(num_test):
# A list of length k storing the labels of the k nearest neighbors to
# the ith test point.
closest_y = []
#########################################################################
# TODO: #
# Use the distance matrix to find the k nearest neighbors of the ith #
# testing point, and use self.y_train to find the labels of these #
# neighbors. Store these labels in closest_y. #
# Hint: Look up the function numpy.argsort. #
#########################################################################
closest_y=self.y_train[np.argsort(dists[i])[:k]]
#########################################################################
# TODO: #
# Now that you have found the labels of the k nearest neighbors, you #
# need to find the most common label in the list closest_y of labels. #
# Store this label in y_pred[i]. Break ties by choosing the smaller #
# label. #
#########################################################################
y_pred[i]=np.argmax(np.bincount(closest_y))
#########################################################################
# END OF YOUR CODE #
#########################################################################
return y_pred
下面是最上面几个算法的理解
1.compute_distances_two_loops 方法
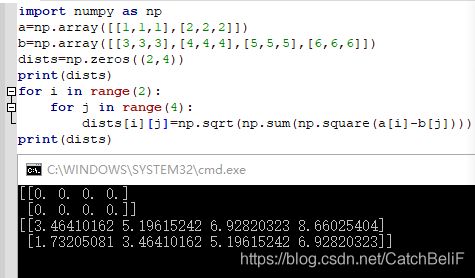
2.compute_distances_one_loops 方法
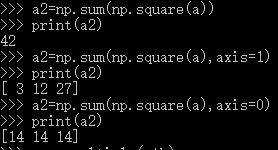
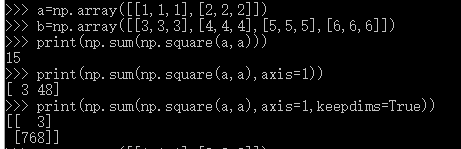
axis=1按行相加
axis=0按列相加
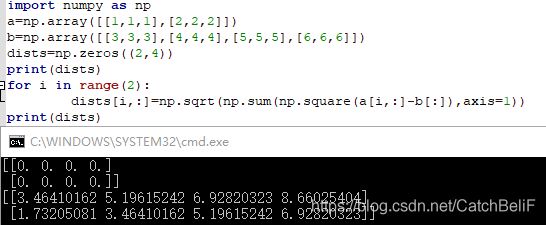
3.compute_distances_no_loops 方法
下图引用而来