白化Whitening
白化操作的目的是让我们的减少冗余信息,准确来说通过白化操作我们有两个目的:
- 每个特征之间关联性更少
- 每个特征有相同的方差
对于第一个目的来说,我们可以通过熟悉的PCA来实现。
PCA
Principal Components Analysis (PCA) 是一个用来减少特征纬度的算法,它通过减少特征的纬度来减少冗余信息。
比如说,下图所示的一个特征纬度为2的点分类问题,我们可以看到数据的主要分布在y=x这条线周围。
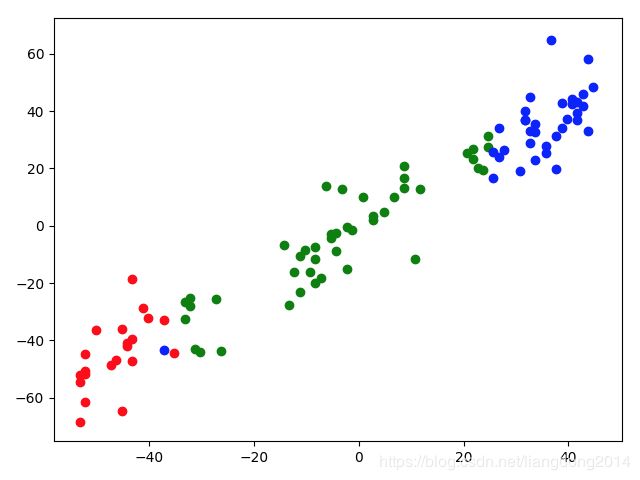
- 我们再回顾一下PCA的计算流程
- 首先计算数据的协方差矩阵 Σ \Sigma Σ
- 接下来对数据进行特征值分解
- 使用原始数据分布乘以特征向量,得到新的特征空间的表示
- 统一期间,我们用 r i r_i ri表示第 i i i个example的向量,它是一个列向量(纬度是 K K K)。 X X X表示整个数据集,shape是 K ∗ N K*N K∗N, N N N是样本的个数
- 如果数据集的均值不在0点,则 X = X − m e a n X X=X-mean_X X=X−meanX将数据的中心点移动到0点。
- 然后计算 Σ \Sigma Σ, Σ = 1 N ∑ i = 0 i = N r i r i T \Sigma = \frac{1}{N}\sum_{i=0}^{i=N}{r_ir_i^T} Σ=N1∑i=0i=NririT,所以 Σ \Sigma Σ的shape是 K ∗ K K*K K∗K
- 然后计算进行特征值分解,将特征向量,按照特征值的大小进行排序。得到 u 1 , u 2 , . . . , u k u_1, u_2, ..., u_k u1,u2,...,uk 以及 λ 1 , λ 2 , . . . , λ k \lambda_1, \lambda_2, ..., \lambda_k λ1,λ2,...,λk,其中 λ 1 > λ 2 > . . . > λ k \lambda_1 > \lambda_2 > ... > \lambda_k λ1>λ2>...>λk
- 至此,我们可以得到一个 r i r_i ri在新空间的表示 r i ′ = ( u 1 T ∗ r i , u 2 T ∗ r i , . . . , u k T ∗ r i ) r_i^{'}=(u_1^T * r_i , u_2^T * r_i, ..., u_k^T * r_i) ri′=(u1T∗ri,u2T∗ri,...,ukT∗ri)。
- 那么怎么从K个纬度中选择出有效的信息呢?我们需要在有效性和效率之间有所平衡。假设说我们规定选择的信息占全部信息的90%即可。那怎么度量这个占比呢?我们假设信息占比标记为 ϵ = ∑ i = 1 K s λ i ∑ i = 1 K λ i \epsilon=\frac{\sum_{i=1}^{K_s}\lambda_i}{\sum_{i=1}^{K}\lambda_i} ϵ=∑i=1Kλi∑i=1Ksλi。其中 K s K_s Ks代表的是我们选择的特征的个数。
- 至此,我们已经可以得到在新的向量空间的特征,并且也可以减少冗余信息。注意⚠️,我们得到的新的特征,每个特征之间是彼此独立的。他们是原有的特征乘以相互正交的向量得到的。
- 在PCA的最后,我们可以可视化一下最后的结果。结果如下所示。可以看到经过处理后,相当于将原来的向量旋转到了和X轴平行的位置。此时X轴的坐标就是在原来坐标空间方差最大方向的坐标。
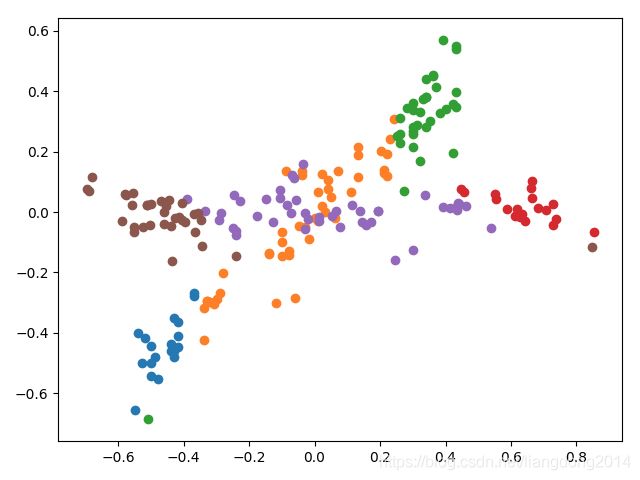
PCA Whitening
- 至此,我们已经可以完成第一个目标了,即就是操作后每个特征之间关联性更少。那么怎么完成第二个目标呢?即就是每个特征有相同的方差。
- PCA whitening的操作方式是 r i w h i t e n i n g = r i P C A λ i r_{i_{whitening}}=\frac{r_{i_{PCA}}}{\lambda_i} riwhitening=λiriPCA就是对PCA计算得到的特征向量的每一纬度再除以一个对应的特征值(表示的就是方差)。除以方差之后,就可以让每个特征值的范围差距不会特别大。
- 接下来给大家展示一下白化后的效果

ZCA whitening
- ZCA whitening是在PCA whitening的基础上再将坐标transform到原来的坐标系,具体的公示如下:
r i Z C A = r i P C A ∗ U T r_{i_{ZCA}} = r_{i_{PCA}} * U^T riZCA=riPCA∗UT
代码
# -*- coding=utf-8 -*-
import numpy as np
import matplotlib.pyplot as plt
from sklearn import datasets
def draw_points(data, label):
color = ['r', 'g', 'b']
for idx in np.unique(label):
# plt.scatter(x=data[np.argwhere(label == idx), 0], y=data[np.argwhere(label == idx), 1], c=color[idx])
plt.scatter(x=data[np.argwhere(label == idx), 0], y=data[np.argwhere(label == idx), 1])
# plt.yticks([-0.5, 0.5])
def load_data():
# digits = datasets.load_digits()
# xs = digits.data
# ys = digits.target
# print(np.shape(xs), np.shape(ys))
xs_1 = np.random.randint(0, 30, [30, 1])
xs_2 = np.random.randint(40, 70, [30, 1])
xs_3 = np.random.randint(75, 100, [40, 1])
xs = np.concatenate([xs_1, xs_2, xs_3], axis=0)
ys = []
for x in xs:
flag = np.random.random()
if flag > 0.5:
ys.append(x + np.random.random() * np.random.randint(30))
else:
ys.append(x - np.random.random() * np.random.randint(30))
# ys = xs + np.random.random([100, 1]) * 5
ys = np.asarray(ys, np.float32)
index_1 = np.argwhere(xs <= 20)
index_2 = np.argwhere(np.logical_and(xs > 20, xs < 80))
index_3 = np.argwhere(xs >= 80)
label = np.asarray(range(100), np.int32)
label[index_1] = 0
label[index_2] = 1
label[index_3] = 2
data = np.asarray(np.concatenate([xs, ys], axis=-1), np.float32)
data[:, 0] /= 10
data[:, 1] /= 100
data -= np.mean(data, axis=0)
print('the shape of data is ', np.shape(data))
return data, label
def PCA(data):
# 首先将中心点的位置调整至原点。即就是均值为0
avg = np.mean(data, axis=0)
data = data - avg
# 计算协方差矩阵
data = np.mat(data)
sigma = data.T * data / len(data)
print('The shape of Sigma is ', np.shape(sigma))
print('The Sigma is ', sigma)
eigenvectors, eigenvalues, _ = np.linalg.svd(sigma)
print('the eigenvalues is ', eigenvalues)
print('the eigenvectors is ', eigenvectors)
data_rot = data * eigenvectors
# draw_points(np.asarray(data_rot, np.float32), label)
return data_rot, eigenvalues, eigenvectors
def PCA_whitening(data, select_k):
data_rot, eigenvalues, eigenvectors = PCA(data)
data_whitening = np.copy(data_rot[:, select_k])
data_whitening[:, 0] /= np.sqrt(eigenvalues[0])
data_whitening[:, 1] /= np.sqrt(eigenvalues[1])
# draw_points(data_whitening, label)
return data_whitening, eigenvalues[:select_k], eigenvectors[:, :select_k]
def ZCA_whitening(data, select_k):
data_whitening_pca, _, eigenvectors = PCA_whitening(data, select_k)
print(np.shape(data_whitening_pca), np.shape(eigenvectors))
data_whitening = data_whitening_pca * eigenvectors.T
print(np.shape(data_whitening))
# draw_points(np.asarray(data_whitening, np.float32), label)
return data_whitening
if __name__ == '__main__':
data, label = load_data()
# draw_points(data, label)
# data_rot, eigenvalues = PCA(data, label)
# draw_points(np.asarray(data_rot, np.float32), label)
# PCA_whitening(data, label)
data_whitening_zca = ZCA_whitening(data, select_k=1)
draw_points(np.asarray(data_whitening_zca, np.float32), label)
plt.show()
参考文献
- whitening