关于医疗影像的mhd和dcm格式图像的读取和坐标转换
本篇博客主要对近年来大赛(Luna16,kaggle,天池)中使用的肺部图像的读取和坐标转换进行整理,如果有错误,欢迎批评指正,谢谢。
1 介绍mhd格式的数据:
数据可以在Luna16(https://luna16.grand-challenge.org/Data/)下载。
(1) 每个病例的数据的存储都是由一个.mhd和一个.raw格式的文件组成。
mhd的内容和比较重要的注释:
ObjectType = Image
NDims = 3 #三维数据
BinaryData = True #二进制数据
BinaryDataByteOrderMSB = False
CompressedData = False
TransformMatrix = 1 0 0 0 1 0 0 0 1 #100,010,001 分别代表x,y,z
Offset = -198.10000600000001 -195 -335.209991 #原点坐标
CenterOfRotation = 0 0 0
AnatomicalOrientation = RAI
ElementSpacing = 0.7617189884185791 0.7617189884185791 2.5 #像素间隔 x,y,z
DimSize = 512 512 121 #数据的大小 x,y,z
ElementType = MET_SHORT
ElementDataFile = 1.3.6.1.4.1.14519.5.2.1.6279.6001.105756658031515062000744821260.raw #数据存储的文件名
(2) mhd已经交代了图像数据的信息,接下来对图像数据进行读取,这里主要用的Python中的SimpleITK库:
以一个病例为例:
import SimpleITK as sitk
import matplotlib.pyplot as plt
case_path = './1.3.6.1.4.1.14519.5.2.1.6279.6001.126264578931778258890371755354.mhd'
itkimage = sitk.ReadImage(case_path) #这部分给出了关于图像的信息,可以打印处理查看,这里就不在显示了
#print(itkimage)
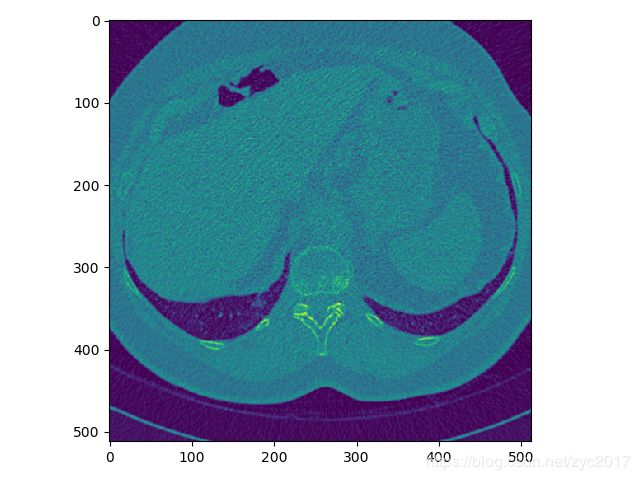
image = sitk.GetArrayFromImage(itkimage) #z,y,x
#查看第100张图像
plt.figure()
plt.imshow(image[100,:,:])
(3) 坐标转换
在luna16大赛中提供了医生标注肺结节位置信息的csv文件 和病例的图像(mhd)。
csv文件中的内容形式为:
seriesuid:表示每个病例图像对应的文件名
coordX,coordX,coordX,diameter_mm:表示医生标注的结节位置信息和直径
在使coordX用卷积网络对肺结节进行检测时,我们需要根据医生提供的标注信息,在图像中找到相应的肺结节位置,接下来说医生标注的坐标与图像中的坐标的关系。
以一个病例上的一个肺结节为例:
csv:
![]()
mhd中给定了图像中的原点坐标为(-163.1962890625, -319.1962890625, -380.5) #x,y,z
像素间隔为(0.607421875 ,0.607421875 ,0.5) #x,y,z
通过以上信息可以计算结节相对原点的坐标,然后用这个坐标除以像素间隔,即为在图像中对应的结节位置。
代码:
#世界坐标转换到图像中的坐标
def worldToVoxelCoord(worldCoord, origin, spacing):
stretchedVoxelCoord = np.absolute(worldCoord - origin)
voxelCoord = stretchedVoxelCoord / spacing
return voxelCoord#图像上的坐标转换为世界坐标:
def VoxelToWorldCoord(voxelCoord, origin, spacing):
strechedVocelCoord = voxelCoord * spacing
worldCoord = strechedVocelCoord + origin
return worldCoord2、介绍dcm格式的数据
数据可以从kaggle(https://www.kaggle.com/c/data-science-bowl-2017/data)或LIDC-IDRC(https://wiki.cancerimagingarchive.net/display/Public/LIDC-IDRI)下载
与mhd文件不同的是,每个病例都是由几十到几百张的切片构成。
接下来以一个病例的一个切片为例:
(1)dcm格式文件的读取:
import dicom
import matplotlib.pyplot as plt
case_path = './1.3.6.1.4.1.14519.5.2.1.6279.6001.179049373636438705059720603192/000001.dcm'
dicomimage = dicom.read_file(case_path) #给出了图像中的信息
print(dicomimage)(0008, 0005) Specific Character Set CS: 'ISO_IR 100'
(0008, 0008) Image Type CS: ['ORIGINAL', 'PRIMARY', 'AXIAL']
(0008, 0016) SOP Class UID UI: CT Image Storage
(0008, 0018) SOP Instance UID UI: 1.3.6.1.4.1.14519.5.2.1.6279.6001.143451261327128179989900675595 #用于唯一区分每一张dcm切片
(0008, 0020) Study Date DA: '20000101'
(0008, 0021) Series Date DA: '20000101'
(0008, 0022) Acquisition Date DA: '20000101'
(0008, 0023) Content Date DA: '20000101'
(0008, 0024) Overlay Date DA: '20000101'
(0008, 0025) Curve Date DA: '20000101'
(0008, 002a) Acquisition DateTime DT: '20000101'
(0008, 0030) Study Time TM: ''
(0008, 0032) Acquisition Time TM: ''
(0008, 0033) Content Time TM: ''
(0008, 0050) Accession Number SH: '2819497684894126'
(0008, 0060) Modality CS: 'CT'
(0008, 0070) Manufacturer LO: 'GE MEDICAL SYSTEMS'
(0008, 0090) Referring Physician's Name PN: ''
(0008, 1090) Manufacturer's Model Name LO: 'LightSpeed Plus'
(0008, 1155) Referenced SOP Instance UID UI: 1.3.6.1.4.1.14519.5.2.1.6279.6001.675906998158803995297223798692
(0010, 0010) Patient's Name PN: ''
(0010, 0020) Patient ID LO: 'LIDC-IDRI-0001'
(0010, 0030) Patient's Birth Date DA: ''
(0010, 0040) Patient's Sex CS: ''
(0010, 1010) Patient's Age AS: ''
(0010, 21d0) Last Menstrual Date DA: '20000101'
(0012, 0062) Patient Identity Removed CS: 'YES'
(0012, 0063) De-identification Method LO: 'DCM:113100/113105/113107/113108/113109/113111'
(0013, 0010) Private Creator LO: 'CTP'
(0013, 1010) Private tag data LO: 'LIDC-IDRI'
(0013, 1013) Private tag data LO: '62796001'
(0018, 0010) Contrast/Bolus Agent LO: 'IV'
(0018, 0015) Body Part Examined CS: 'CHEST'
(0018, 0022) Scan Options CS: 'HELICAL MODE'
(0018, 0050) Slice Thickness DS: '2.500000' #切片的厚度,注:不同仪器,会导致不同的切片厚度,但是同一个病例上的切片厚度是相同的
(0018, 0060) KVP DS: '120'
(0018, 0090) Data Collection Diameter DS: '500.000000'
(0018, 1020) Software Version(s) LO: 'LightSpeedApps2.4.2_H2.4M5'
(0018, 1100) Reconstruction Diameter DS: '360.000000'
(0018, 1110) Distance Source to Detector DS: '949.075012'
(0018, 1111) Distance Source to Patient DS: '541.000000'
(0018, 1120) Gantry/Detector Tilt DS: '0.000000'
(0018, 1130) Table Height DS: '144.399994'
(0018, 1140) Rotation Direction CS: 'CW'
(0018, 1150) Exposure Time IS: '570'
(0018, 1151) X-Ray Tube Current IS: '400'
(0018, 1152) Exposure IS: '4684'
(0018, 1160) Filter Type SH: 'BODY FILTER'
(0018, 1170) Generator Power IS: '48000'
(0018, 1190) Focal Spot(s) DS: '1.200000'
(0018, 1210) Convolution Kernel SH: 'STANDARD'
(0018, 5100) Patient Position CS: 'FFS'
(0020, 000d) Study Instance UID UI: 1.3.6.1.4.1.14519.5.2.1.6279.6001.298806137288633453246975630178 # 每个病例对应的检查实例号
(0020, 000e) Series Instance UID UI: 1.3.6.1.4.1.14519.5.2.1.6279.6001.179049373636438705059720603192 #不同检查对应的序列实例号, 注:比如一个病例中包括不同形式切片,像x光和对肺部不同显示的CT切片,如果想要对他们进行归类时,可以使用这个实例号
(0020, 0010) Study ID SH: ''
(0020, 0011) Series Number IS: '3000566'
(0020, 0013) Instance Number IS: '80'
(0020, 0032) Image Position (Patient) DS: ['-166.000000', '-171.699997', '-207.500000'] #表示图像的左上角在空间坐标系中的x,y,z坐标,单位是毫米
(0020, 0037) Image Orientation (Patient) DS: ['1.000000', '0.000000', '0.000000', '0.000000', '1.000000', '0.000000']
(0020, 0052) Frame of Reference UID UI: 1.3.6.1.4.1.14519.5.2.1.6279.6001.229925374658226729607867499499
(0020, 1040) Position Reference Indicator LO: 'SN'
(0020, 1041) Slice Location DS: '-207.500000' #为切片z轴相对位置
(0028, 0002) Samples per Pixel US: 1
(0028, 0004) Photometric Interpretation CS: 'MONOCHROME2'
(0028, 0010) Rows US: 512
(0028, 0011) Columns US: 512
(0028, 0030) Pixel Spacing DS: ['0.703125', '0.703125'] #像素间隔
(0028, 0100) Bits Allocated US: 16
(0028, 0101) Bits Stored US: 16
(0028, 0102) High Bit US: 15
(0028, 0103) Pixel Representation US: 1
(0028, 0120) Pixel Padding Value US: 63536
(0028, 0303) Longitudinal Temporal Information M CS: 'MODIFIED'
(0028, 1050) Window Center DS: '-600'
(0028, 1051) Window Width DS: '1600'
(0028, 1052) Rescale Intercept DS: '-1024' #有的CT图像中像素不是HU值,Rescale Intercept和Rescale Slope这两个用于将像素转化为HU值
(0028, 1053) Rescale Slope DS: '1'
(0038, 0020) Admitting Date DA: '20000101'
(0040, 0002) Scheduled Procedure Step Start Date DA: '20000101'
(0040, 0004) Scheduled Procedure Step End Date DA: '20000101'
(0040, 0244) Performed Procedure Step Start Date DA: '20000101'
(0040, 2016) Placer Order Number / Imaging Servi LO: ''
(0040, 2017) Filler Order Number / Imaging Servi LO: ''
(0040, a075) Verifying Observer Name PN: 'Removed by CTP'
(0040, a123) Person Name PN: 'Removed by CTP'
(0040, a124) UID UI: 1.3.6.1.4.1.14519.5.2.1.6279.6001.335419887712224178340067932923
(0070, 0084) Content Creator's Name PN: ''
(0088, 0140) Storage Media File-set UID UI: 1.3.6.1.4.1.14519.5.2.1.6279.6001.211790042620307056609660772296
(7fe0, 0010) Pixel Data OW: Array of 524288 bytes
以上我对一些重要的信息做了标注。
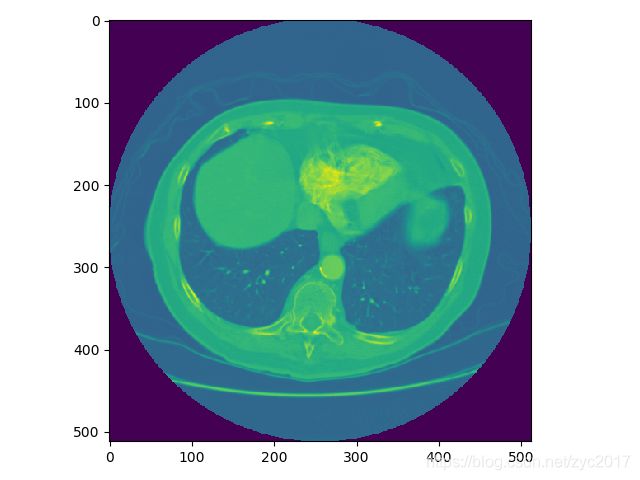
CT图像可视化:
image= dicomimage.pixel_array
plt.figure(2)
plt.imshow(image)(2)坐标转换和mhd类似。
(3)接下来直接贴比较重要的代码了:
a、将一个病例的所有图像进行堆叠:
slices = [dicom.read_file(s) for s in dcm]
slices.sort(key=lambda x: float(x.ImagePositionPatient[2])) #从小到大排序 从肺的底部到头部 #x.ImagePositionPatient[2]表示切片空间对应的位置,即z轴b、将CT图像中的像素转换成HU值
for slice_number in range(len(slices)): #slices表示一个病例的所有切片
intercept = slices[slice_number].RescaleIntercept
slope = slices[slice_number].RescaleSlope
if slope != 1:
image[slice_number] = slope * image[slice_number].astype(np.float64)
image[slice_number] = image[slice_number].astype(np.int16)
image[slice_number] += np.int16(intercept)