Author: Zuguang Gu ( [email protected] )
翻译:诗翔
Date: 2018-10-30
热图列表可以提高多种数据资源关系的可视化。在这个手册中,我们将讨论创建热图列表的配置。你可以在Examples 文档和 文章附加文档 中找到真实案例。
热图串联
你可以在图形中从左到右排列多个热图。实际上,一个单一的热图是热图列表长度为1时的特殊情况。
Heatmap()实际上是单一热图的类构造函数。如果需要组合超过一个热图,用户可以通过+操作符添加热图。
library(ComplexHeatmap)
mat1 = matrix(rnorm(80, 2), 8, 10)
mat1 = rbind(mat1, matrix(rnorm(40, -2), 4, 10))
rownames(mat1) = paste0("R", 1:12)
colnames(mat1) = paste0("C", 1:10)
mat2 = matrix(rnorm(60, 2), 6, 10)
mat2 = rbind(mat2, matrix(rnorm(60, -2), 6, 10))
rownames(mat2) = paste0("R", 1:12)
colnames(mat2) = paste0("C", 1:10)
ht1 = Heatmap(mat1, name = "ht1")
ht2 = Heatmap(mat2, name = "ht2")
class(ht1)
## [1] "Heatmap"
## attr(,"package")
## [1] "ComplexHeatmap"
class(ht2)
## [1] "Heatmap"
## attr(,"package")
## [1] "ComplexHeatmap"
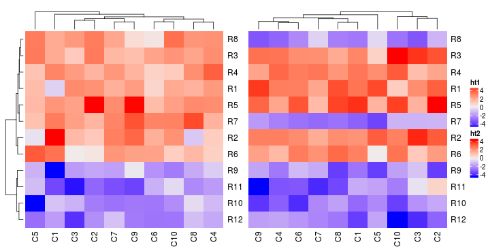
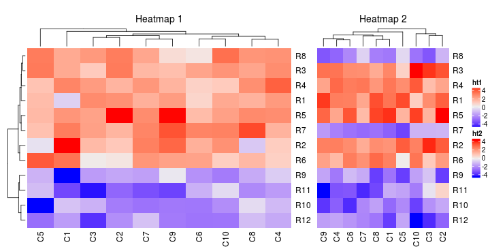
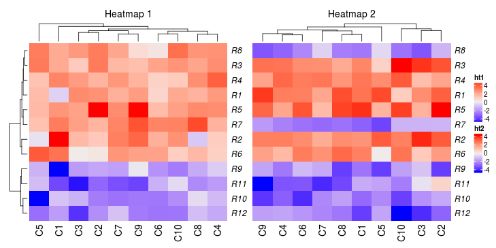
ht1 + ht2
在默认情况下,第二个热图的行树状图会被移除,行的顺序会和第一个热图保持一致。
两个热图相加的返回值是一个HeatmapList对象。直接允许ht_list对象会默认调用draw()方法。通过显式地调用draw()方法,你可以进行更多的控制,例如图例和标题。
ht_list = ht1 + ht2
class(ht_list)
## [1] "HeatmapList"
## attr(,"package")
## [1] "ComplexHeatmap"
你可以添加任意数目到热图到一个热图列表。你也可以将一个热图列表添加到一个热图列表。
ht1 + ht1 + ht1
ht1 + ht_list
ht_list + ht1
ht_list + ht_list
NULL也可以被添加到一个热图列表。如果用户想要通过for循环构造一个热图列表,这会非常地方便。
ht_list = NULL
for(s in sth) {
ht_list = ht_list + Heatmap(...)
}
标题
热图列表的标题独立于热图的标题。
ht1 = Heatmap(mat1, name = "ht1", row_title = "Heatmap 1", column_title = "Heatmap 1")
ht2 = Heatmap(mat2, name = "ht2", row_title = "Heatmap 2", column_title = "Heatmap 2")
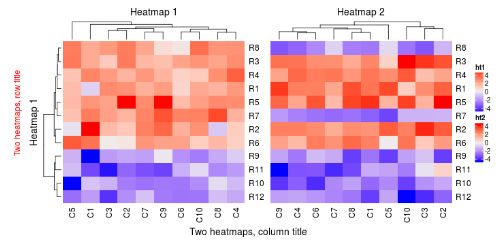
ht_list = ht1 + ht2
draw(ht_list, row_title = "Two heatmaps, row title", row_title_gp = gpar(col = "red"),
column_title = "Two heatmaps, column title", column_title_side = "bottom")
热图间隔
热图间隔可以使用一个unit对象传入gap参数。
draw(ht_list, gap = unit(1, "cm"))
draw(ht_list + ht_list, gap = unit(c(3, 6, 9, 0), "mm"))
热图大小
热图的宽度可以设置为固定长度。
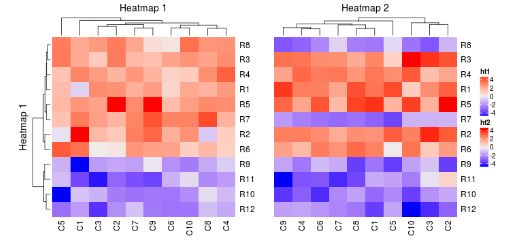
ht1 = Heatmap(mat1, name = "ht1", column_title = "Heatmap 1")
ht2 = Heatmap(mat2, name = "ht2", column_title = "Heatmap 2", width = unit(5, "cm"))
ht1 + ht2
或者宽度可以设置为相对值。注意在这种情况下,每个热图都需要进行设置。
ht1 = Heatmap(mat1, name = "ht1", column_title = "Heatmap 1", width = 2)
ht2 = Heatmap(mat2, name = "ht2", column_title = "Heatmap 2", width = 1)
ht1 + ht2
自动调整
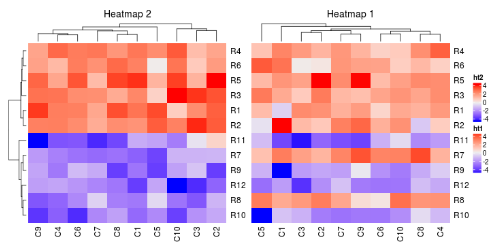
如果绘制超过一个热图,会进行一些自动的调整。默认第一个热图是主热图。其他热图会根据主热图的设定进行更改。调整包括:
- 行聚类被移除
- 行标题被移除
- 如果主热图按行切分,余下的热图也会进行该操作
主热图可以通过main_heatmap参数指定。值可以是一个数值索引或者热图的名字。
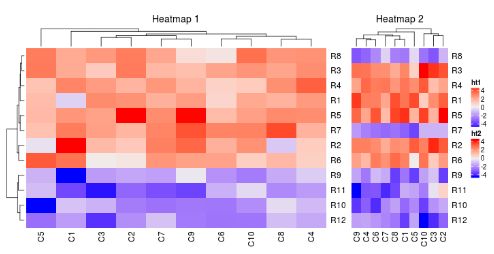
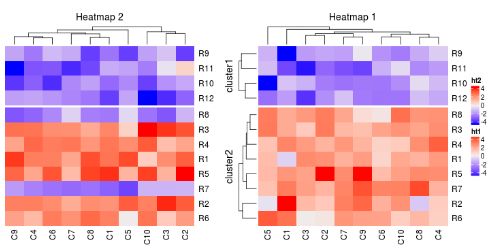
ht1 = Heatmap(mat1, name = "ht1", column_title = "Heatmap 1", km = 2)
ht2 = Heatmap(mat2, name = "ht2", column_title = "Heatmap 2")
ht1 + ht2
# note we changed the order of `ht1` and `ht2`
draw(ht2 + ht1)
# here although `ht1` is the second heatmap, we specify `ht1` to be
# the main heatmap by explicitely setting `main_heatmap` argument
draw(ht2 + ht1, main_heatmap = "ht1")
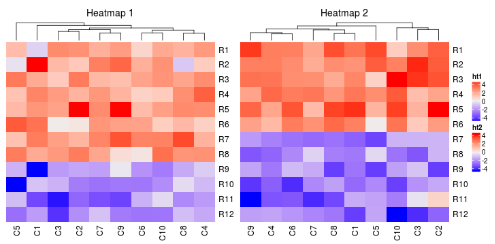
如果主热图没有行聚类,其他热图也不会有。
ht1 = Heatmap(mat1, name = "ht1", column_title = "Heatmap 1", cluster_rows = FALSE)
ht2 = Heatmap(mat2, name = "ht2", column_title = "Heatmap 2")
ht1 + ht2
同时更改图形参数
ht_global_opt()可以设置维度名和标题名图形参数为全局设定。
ht_global_opt(heatmap_row_names_gp = gpar(fontface = "italic"),
heatmap_column_names_gp = gpar(fontsize = 14))
ht1 = Heatmap(mat1, name = "ht1", column_title = "Heatmap 1")
ht2 = Heatmap(mat2, name = "ht2", column_title = "Heatmap 2")
ht1 + ht2
ht_global_opt(RESET = TRUE)
下面是支持的所有选项,你也可以控制图例设置。
names(ht_global_opt())
## [1] "heatmap_row_names_gp" "heatmap_column_names_gp"
## [3] "heatmap_row_title_gp" "heatmap_column_title_gp"
## [5] "heatmap_legend_title_gp" "heatmap_legend_title_position"
## [7] "heatmap_legend_labels_gp" "heatmap_legend_grid_height"
## [9] "heatmap_legend_grid_width" "heatmap_legend_grid_border"
## [11] "annotation_legend_title_gp" "annotation_legend_title_position"
## [13] "annotation_legend_labels_gp" "annotation_legend_grid_height"
## [15] "annotation_legend_grid_width" "annotation_legend_grid_border"
## [17] "fast_hclust"
获取顺序和树状图
row_order,column_order, row_dend 和 column_dend可以用来从热图中获取对应的信息。下面例子展示了其用法非常直观。
ht_list = ht1 + ht2
row_order(ht_list)
## [[1]]
## [1] 8 3 4 1 5 7 2 6 9 11 10 12
column_order(ht_list)
## $ht1
## [1] 5 1 3 2 7 9 6 10 8 4
##
## $ht2
## [1] 9 4 6 7 8 1 5 10 3 2
row_dend(ht_list)
## [[1]]
## 'dendrogram' with 2 branches and 12 members total, at height 16.14288
column_dend(ht_list)
## $ht1
## 'dendrogram' with 2 branches and 10 members total, at height 8.069474
##
## $ht2
## 'dendrogram' with 2 branches and 10 members total, at height 6.883
如果ht_list还没有绘制,如果矩阵很大使用这4个函数会有点慢。但是如果ht_list已经绘制了,这意味着已经对矩阵进行了聚类,那这些函数就会很快。
ht_list = draw(ht1 + ht2)
row_order(ht_list)
column_order(ht_list)
row_dend(ht_list)
column_dend(ht_list)
带行注释的热图列表
请查看 Heatmap Annotation 获取信息。
修改主热图的行顺序/聚类
这可以直接调用draw()方法实现。
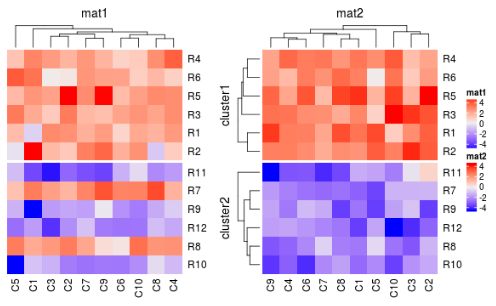
split = rep(c("a", "b"), each = 6)
ht_list = Heatmap(mat1, name = "mat1", cluster_rows = FALSE, column_title = "mat1") +
Heatmap(mat2, name = "mat2", cluster_rows = FALSE, column_title = "mat2")
draw(ht_list, main_heatmap = "mat1", split = split)
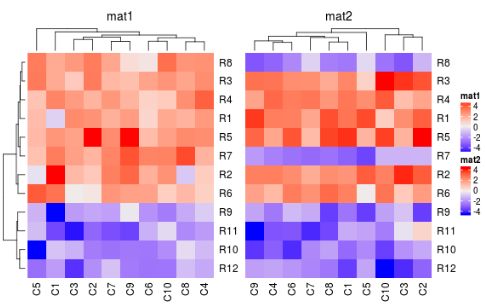
draw(ht_list, main_heatmap = "mat2", km = 2, cluster_rows = TRUE)
draw(ht_list, cluster_rows = TRUE, main_heatmap = "mat1", show_row_dend =TRUE)
draw(ht_list, cluster_rows = TRUE, main_heatmap = "mat2", show_row_dend =TRUE)
会话信息
sessionInfo()
## R version 3.5.1 Patched (2018-07-12 r74967)
## Platform: x86_64-pc-linux-gnu (64-bit)
## Running under: Ubuntu 16.04.5 LTS
##
## Matrix products: default
## BLAS: /home/biocbuild/bbs-3.8-bioc/R/lib/libRblas.so
## LAPACK: /home/biocbuild/bbs-3.8-bioc/R/lib/libRlapack.so
##
## locale:
## [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C LC_TIME=en_US.UTF-8
## [4] LC_COLLATE=C LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
## [7] LC_PAPER=en_US.UTF-8 LC_NAME=C LC_ADDRESS=C
## [10] LC_TELEPHONE=C LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
##
## attached base packages:
## [1] stats4 parallel grid stats graphics grDevices utils datasets methods
## [10] base
##
## other attached packages:
## [1] dendextend_1.9.0 dendsort_0.3.3 cluster_2.0.7-1 IRanges_2.16.0
## [5] S4Vectors_0.20.0 BiocGenerics_0.28.0 HilbertCurve_1.12.0 circlize_0.4.4
## [9] ComplexHeatmap_1.20.0 knitr_1.20 markdown_0.8
##
## loaded via a namespace (and not attached):
## [1] mclust_5.4.1 Rcpp_0.12.19 mvtnorm_1.0-8 lattice_0.20-35
## [5] png_0.1-7 class_7.3-14 assertthat_0.2.0 mime_0.6
## [9] R6_2.3.0 GenomeInfoDb_1.18.0 plyr_1.8.4 evaluate_0.12
## [13] ggplot2_3.1.0 highr_0.7 pillar_1.3.0 GlobalOptions_0.1.0
## [17] zlibbioc_1.28.0 rlang_0.3.0.1 lazyeval_0.2.1 diptest_0.75-7
## [21] kernlab_0.9-27 whisker_0.3-2 GetoptLong_0.1.7 stringr_1.3.1
## [25] RCurl_1.95-4.11 munsell_0.5.0 compiler_3.5.1 pkgconfig_2.0.2
## [29] shape_1.4.4 nnet_7.3-12 tidyselect_0.2.5 gridExtra_2.3
## [33] tibble_1.4.2 GenomeInfoDbData_1.2.0 viridisLite_0.3.0 crayon_1.3.4
## [37] dplyr_0.7.7 MASS_7.3-51 bitops_1.0-6 gtable_0.2.0
## [41] magrittr_1.5 scales_1.0.0 stringi_1.2.4 XVector_0.22.0
## [45] viridis_0.5.1 flexmix_2.3-14 bindrcpp_0.2.2 robustbase_0.93-3
## [49] fastcluster_1.1.25 HilbertVis_1.40.0 rjson_0.2.20 RColorBrewer_1.1-2
## [53] tools_3.5.1 fpc_2.1-11.1 glue_1.3.0 trimcluster_0.1-2.1
## [57] DEoptimR_1.0-8 purrr_0.2.5 colorspace_1.3-2 GenomicRanges_1.34.0
## [61] prabclus_2.2-6 bindr_0.1.1 modeltools_0.2-22