数据分析整理,R,caret包(1)
caret包可以处理至少以下事情.
1、初步筛选属性(过滤以下属性)
a、找出 属性值接近为常数的 属性 nearZeroVar
b、找出 相关系数最大的 属性 findCorrelation
c、找出 多重共线性的 属性 findLinearCombos
2、处理缺失值
preProcess(data, method=c("bagImpute","knnImpute"));predict(pro, newdata)
3、中心化、标准化
preProcess(data, method=c("center","scale"))
4、特征选择
rfeControl,rfe
sbfControl,sbf
5、抽样数据划分
createDataPartition()
createFold()…
6、模型训练
trainControl():设置训练交叉验证的重数,重复几次等
train(): 设置使用何种模型训练(查看函数定义[非常之多])
7、预测结果
predict()
library(C50)
library(lattice)library(ggplot2)
library(caret)
#多药耐药逆转剂
#528 obs. of 342 variables
#528个化合物,342个描述符
data(mdrr)
# 0 variance
newdata <- mdrrDescr[, -nearZeroVar(mdrrDescr)]# high cor
descrCorr <- cor(newdata)
newdata2 <- newdata[, -findCorrelation(descrCorr)]
# 去掉共线性(如果存在)
comboInfo <- findLinearCombos(newdata2)
if(!is.null(comboInfo)){
newdata3 <- newdata2[, -comboInfo$remove]
}
# 如果有缺失值,使用bagImpute,knnImpute进行计算填补
if(nrow(newdata2[!complete.cases(newdata2),])!=0)
{
process <- preProcess(newdata2, method="bagImpute")
pre <- predict(process, newdata2)
}
# feather selection
# 产生检测属性个数的序列
subsets <- seq(2, ncol(newdata2), by=2)
# define sbfControl
sbfControls_rf <- sbfControl( functions = rfSBF, method = 'cv', repeats = 5)
pro <- sbf(newdata2, mdrrClass, sizes = subsets, sbfControl=sbfControls_rf)
summary(pro)
# feature selected variables
pro$optVariables
# 训练模型
# 获取特征选择后的属性
newdata4 <- newdata2[, pro$optVariables]
# 训练数据和测试数据
index <- createDataPartition(mdrrClass, p=3/4, list=F)
trainx <- newdata4[index,]
trainy <- mdrrClass[index]
testx <- newdata4[-index,]
testy <- mdrrClass[-index]
# 设置模型训练参数并拟合模型
fitControl <- trainControl(method="repeatedcv", number=10, repeats=3, returnResamp="all")
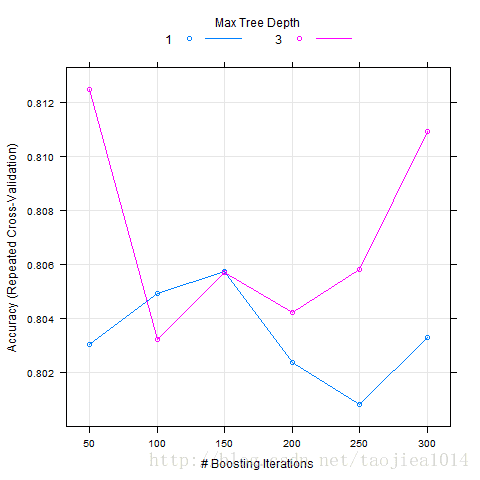
gbmGrid <- expand.grid(interaction.depth=c(1,3), n.trees=seq(50,300,by=50), shrinkage=0.1,n.minobsinnode = 20)
gbmFit1 <- train(trainx, trainy, method="gbm", trControl=fitControl, tuneGrid= gbmGrid, verbose=F)
trainControl
png("foo.png",family="GB1")
plot(gbmFit1)
dev.off()
# 使用训练好的模型进行predict
predict(gbmFit1, newdata=testx)
# 混淆矩阵查看结果
table(testy, predict(gbmFit1, newdata=testx))
testy Active Inactive
Active 61 13
Inactive 12 45
# 使用另外的模型(装袋法)
gbmFit2 <- train(trainx, trainy, method="treebag", trControl=fitControl)
table(testy, predict(gbmFit2, newdata=testx))
testy Active Inactive
Active 64 10
Inactive 13 44
predValues <- extractPrediction(models, testX=testx, testY=testy)
# predValues <- extractPrediction(models, testX=testx)
obs pred model dataType object
1 Active Active gbm Training Object1
2 Active Active gbm Training Object1
3 Inactive Inactive gbm Training Object1
4 Active Inactive gbm Training Object1
5 Active Active gbm Training Object1
6 Active Active gbm Training Object1