Python中的WFDB库使用
Python中的WFDB库使用
- Demo1--使用'rdsamp'函数读取数据并画图
- Demo2--读取ECG信号和标记并画图
- Demo3--仅仅读取头文件
- Demo4--下载数据
- 1:下载整个数据库
- 2:下载数据库中的特定文件
- Demo5--ECG信号处理
- 1:使用GQRS算法定位QRS波位置以及位置的矫正
- 2:使用XQRS检测算法,并将结果与参考注释进行比较
- 正确R波标记位置读取
- 持续更新ing
- 参考文档:
读取的库的内容主要是MITDB和NSTDB(噪音压力测试库),均是在线读取数据
Physionet ATM:https://physionet.org/cgi-bin/atm/ATM
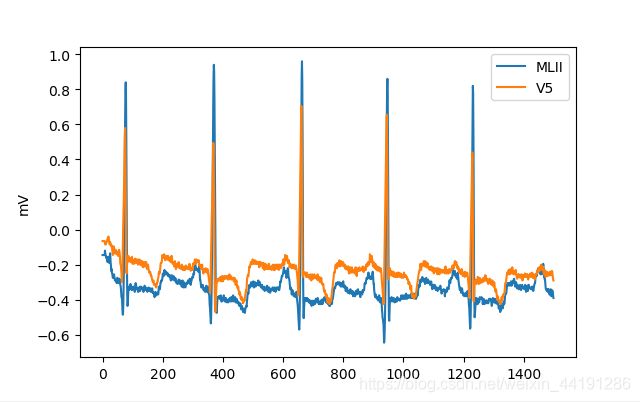
Demo1–使用’rdsamp’函数读取数据并画图
import wfdb
import matplotlib.pyplot as plt
signal, fields=wfdb.rdsamp('100',channels=[0, 1], sampfrom=0, sampto=1500, pb_dir='mitdb/')
#mitdb数据库的是数据是两导联,格式是[650000,2]的数据,channels是选择读取哪一个导联的
print('signal:',signal)
print('fields:',fields)
plt.plot(signal)
plt.ylabel(fields['units'][0])
plt.legend([fields['sig_name'][0],fields['sig_name'][1]])
plt.show()
输出:
signal: [[-0.145 -0.065]
[-0.145 -0.065]
[-0.145 -0.065]
...,
[-0.355 -0.27 ]
[-0.38 -0.285]
[-0.39 -0.29 ]]
fields: {'fs': 360, 'sig_len': 1500, 'n_sig': 2, 'base_date': None, 'base_time': None, 'units': ['mV', 'mV'], 'sig_name': ['MLII', 'V5'], 'comments': ['69 M 1085 1629 x1', 'Aldomet, Inderal']}
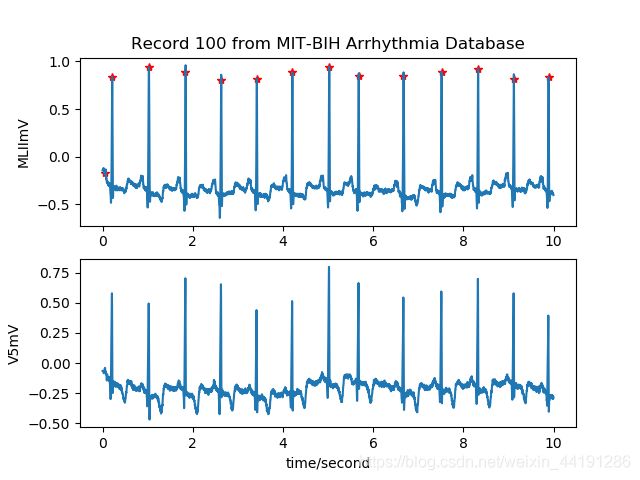
Demo2–读取ECG信号和标记并画图
import wfdb
record=wfdb.rdrecord('100', sampto=3600, pb_dir='mitdb/')
annotation=wfdb.rdann('100', 'atr',sampto=3600, pb_dir='mitdb/')
wfdb.plot_wfdb(record=record, annotation=annotation,
title='Record 100 from MIT-BIH Arrhythmia Database', time_units='seconds')
print('annotation:',annotation.__dict__)
print('record:',record.__dict__)
print('signal:',record.p_signal)#这个record其实并不是字典需要用点操作符取出值
输出:
annotation {'record_name': '100', 'extension': 'atr', 'sample': array([ 18, 77, 370, 662, 946, 1231, 1515, 1809, 2044, 2402, 2706,
2998, 3282, 3560]), 'symbol': ['+', 'N', 'N', 'N', 'N', 'N', 'N', 'N', 'A', 'N', 'N', 'N', 'N', 'N'], 'subtype': array([0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0]), 'chan': array([0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0]), 'num': array([0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0]), 'aux_note': ['(N\x00', '', '', '', '', '', '', '', '', '', '', '', '', ''], 'fs': 360, 'label_store': None, 'description': None, 'custom_labels': None, 'contained_labels': None, 'ann_len': 14}
record: {'record_name': '100', 'n_sig': 2, 'fs': 360, 'counter_freq': None, 'base_counter': None, 'sig_len': 3600, 'base_time': None, 'base_date': None, 'comments': ['69 M 1085 1629 x1', 'Aldomet, Inderal'], 'sig_name': ['MLII', 'V5'], 'p_signal': array([[-0.145, -0.065],
[-0.145, -0.065],
[-0.145, -0.065],
...,
[-0.39 , -0.295],
[-0.4 , -0.29 ],
[-0.405, -0.285]]), 'd_signal': None, 'e_p_signal': None, 'e_d_signal': None, 'file_name': ['100.dat', '100.dat'], 'fmt': ['212', '212'], 'samps_per_frame': [1, 1], 'skew': [None, None], 'byte_offset': [None, None], 'adc_gain': [200.0, 200.0], 'baseline': [1024, 1024], 'units': ['mV', 'mV'], 'adc_res': [11, 11], 'adc_zero': [1024, 1024], 'init_value': [995, 1011], 'checksum': [48184, 1171], 'block_size': [0, 0]}
signal: [[-0.145 -0.065]
[-0.145 -0.065]
[-0.145 -0.065]
...
[-0.39 -0.295]
[-0.4 -0.29 ]
[-0.405 -0.285]]
Demo3–仅仅读取头文件
record=wfdb.rdheader('100', pb_dir='mitdb/')
print(record.__dict__)
输出:
{'record_name': '100', 'n_sig': 2, 'fs': 360, 'counter_freq': None, 'base_counter': None, 'sig_len': 650000, 'base_time': None, 'base_date': None, 'comments': ['69 M 1085 1629 x1', 'Aldomet, Inderal'], 'sig_name': ['MLII', 'V5'], 'p_signal': None, 'd_signal': None, 'e_p_signal': None, 'e_d_signal': None, 'file_name': ['100.dat', '100.dat'], 'fmt': ['212', '212'], 'samps_per_frame': [1, 1], 'skew': [None, None], 'byte_offset': [None, None], 'adc_gain': [200.0, 200.0], 'baseline': [1024, 1024], 'units': ['mV', 'mV'], 'adc_res': [11, 11], 'adc_zero': [1024, 1024], 'init_value': [995, 1011], 'checksum': [-22131, 20052], 'block_size': [0, 0]}
Demo4–下载数据
1:下载整个数据库
import wfdb
import os
from IPython.display import display
dl_dir="C:\\Users\\jinzh\\OneDrive\\python\\Daily_Practice\\2019\\temp_dir"
if __name__ == '__main__': #这一段必须要加
wfdb.dl_database('ahadb', dl_dir)
display(os.listdir(dl_dir))
#wfdb.io.dl_database(db_dir, dl_dir, records='all', annotators='all', keep_subdirs=True, overwrite=False)
输出:
Downloading files...
Finished downloading files
['0001.atr', '0001.dat', '0001.hea', '0201.atr', '0201.dat', '0201.hea', 'data', 'STAFF-Studies-bibliography-2016.pdf']
注:代码中的**if __ name __ == ‘__main __’:**必须要加,否则会产生以下报错,这其实是python进程池multiprocessing.Pool运行错误
RuntimeError:
An attempt has been made to start a new process before the
current process has finished its bootstrapping phase.
This probably means that you are not using fork to start your
child processes and you have forgotten to use the proper idiom
in the main module:
if __name__ == '__main__':
freeze_support()
...
The "freeze_support()" line can be omitted if the program
is not going to be frozen to produce an executable.
2:下载数据库中的特定文件
import wfdb
import os
from IPython.display import display
file_list=['100.hea','100.atr','100.dat']
dl_dir="C:\\Users\\jinzh\\OneDrive\\python\\Daily_Practice\\2019\\temp_dir"
if __name__ == '__main__':
wfdb.dl_files('mitdb', dl_dir, file_list)
display(os.listdir(dl_dir))
#wfdb.io.dl_files(db, dl_dir, files, keep_subdirs=True, overwrite=False)
输出:
Downloading files...
Finished downloading files
['100.atr', '100.dat', '100.hea']
Demo5–ECG信号处理
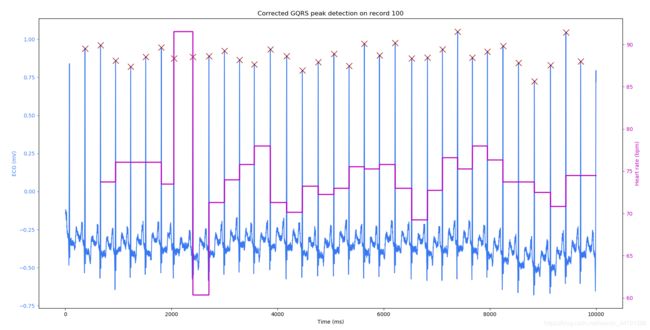
1:使用GQRS算法定位QRS波位置以及位置的矫正
import matplotlib.pyplot as plt
import wfdb
from wfdb import processing
#使用gqrs定位算法矫正峰值位置
def peaks_hr(sig, peak_inds, fs, title, figsize=(20,10), saveto=None):
#这个函数是用来画出信号峰值和心律
#计算心律
hrs=processing.compute_hr(sig_len=sig.shape[0], qrs_inds=peak_inds, fs=fs)
N=sig.shape[0]
fig, ax_left=plt.subplots(figsize=figsize)
ax_right=ax_left.twinx()
ax_left.plot(sig, color='#3979f0', label='Signal')
ax_left.plot(peak_inds, sig[peak_inds], 'rx', marker='x', color='#8b0000', label='Peak', markersize=12)#画出标记
ax_right.plot(np.arange(N), hrs, label='Heart rate', color='m', linewidth=2)#画出心律,y轴在右边
ax_left.set_title(title)
ax_left.set_xlabel('Time (ms)')
ax_left.set_ylabel('ECG (mV)', color='#3979f0')
ax_right.set_ylabel('Heart rate (bpm)', color='m')
#设置颜色使得和线条颜色一致
ax_left.tick_params('y', colors='#3979f0')
ax_right.tick_params('y', colors='m')
if saveto is not None:
plt.savefig(saveto, dpi=600)
plt.show()
#加载ECG信号
record=wfdb.rdrecord('100', sampfrom=0, sampto=10000, channels=[0], pb_dir='mitdb/')
#使用gqrs算法定位qrs波位置
qrs_inds=processing.gqrs_detect(sig=record.p_signal[:, 0], fs=record.fs)
#画出结果
peaks_hr(sig=record.p_signal, peak_inds=qrs_inds, fs=record.fs, title='GQRS peak detection on record 100')
#修正峰值,将其设置为局部最大值
min_bpm=20
max_bpm=230
#使用可能最大的bpm作为搜索半径
search_radius=int(record.fs*60/max_bpm)
corrected_peak_inds=processing.correct_peaks(record.p_signal[:, 0], peak_inds=qrs_inds, search_radius=search_radius, smooth_window_size=150)
#输出结果
print('Uncorrected gqrs detected peak indeices:',sorted(qrs_inds))
print('Corrected gqrs detected peak indices:', sorted(corrected_peak_inds))
peaks_hr(sig=record.p_signal, peak_inds=sorted(corrected_peak_inds), fs=record.fs, title='Corrected GQRS peak detection on record 100')
输出:
Uncorrected gqrs detected peak indeices: [357, 650, 934, 1219, 1502, 1797, 2032, 2390, 2693, 2985, 3270, 3547, 3850, 4158, 4453, 4752, 5048, 5334, 5621, 5906, 6202, 6514, 6811, 7093, 7379, 7657, 7940, 8233, 8526, 8825, 9129, 9419, 9698]
Corrected gqrs detected peak indices: [370, 663, 947, 1231, 1515, 1809, 2045, 2403, 2706, 2998, 3283, 3560, 3863, 4171, 4466, 4765, 5061, 5347, 5634, 5919, 6215, 6527, 6824, 7106, 7393, 7670, 7953, 8246, 8539, 8837, 9142, 9432, 9710]
2:使用XQRS检测算法,并将结果与参考注释进行比较
sig, fields=wfdb.rdsamp('100', channels=[0], sampto=15000, pb_dir='mitdb/')
ann_ref=wfdb.rdann('100', 'atr', sampto=15000, pb_dir='mitdb/')
#使用XQRS算法
xqrs=processing.XQRS(sig=sig[:,0], fs=fields['fs'])
xqrs.detect()
#这里还可以直接使用xqrs_detection
#qrs_inds=processing.xqrs_detect(sig=sig[:,0], fs=fields['fs'])
#下面进行算法的结果和注释中的结果相对比
#注意:在100.atr中的qrs注释第一个数是18,而18这个位置上并没有峰值,真正的第一个峰值是在第二个数77开始的所以是[1:]
comparitor=processing.compare_annotations(ref_sample=ann_ref.sample[1:],#见上面注释
test_sample=xqrs.qrs_inds,
window_width=int(0.1*fields['fs']),
signal=sig[:,0])
#输出结果
comparitor.print_summary()
fig=comparitor.plot(title='XQRS detected QRS vs reference annotations',return_fig=True)
# display(fig[0])
plt.show(fig[0])#这一步必须加,不然图片会一闪而逝
输出:
Learning initial signal parameters...
Found 8 beats during learning. Initializing using learned parameters
Running QRS detection...
QRS detection complete.
51 reference annotations, 51 test annotations
True Positives (matched samples): 51
False Positives (unmatched test samples: 0
False Negatives (unmatched reference samples): 0
Specificity: 1.0000 (51/51)
Positive Predictivity: 1.0000 (51/51)
False Positive Rate: 0.0000 (0/51)
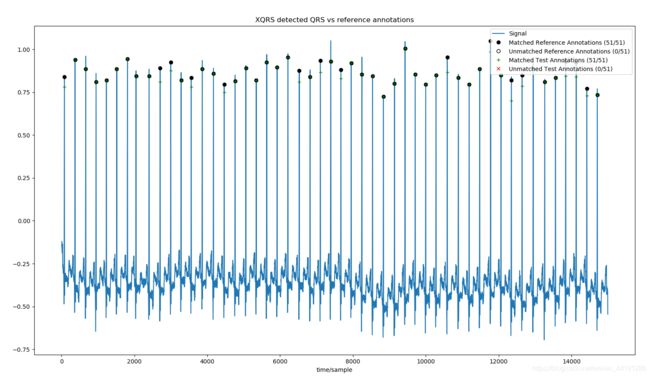
注:如果直接和官方文档里一样使用comparitor.plot(title=‘XQRS detected QRS vs reference annotations’)来输出图像会一闪而逝,需要如上述代码一样修改以下,这个plot函数还有其他的参数设置,具体参考如下:
plot(sig_style='', title=None, figsize=None, return_fig=False)
Plot the comparison of two sets of annotations, possibly overlaid on their original signal.
sig_style : str, optional
The matplotlib style of the signal
title : str, optional
The title of the plot
figsize: tuple, optional
Tuple pair specifying the width, and height of the figure. It is the’figsize’ argument passed into matplotlib.pyplot’s figure function.
return_fig : bool, optional
Whether the figure is to be returned as an output argument.
正确R波标记位置读取
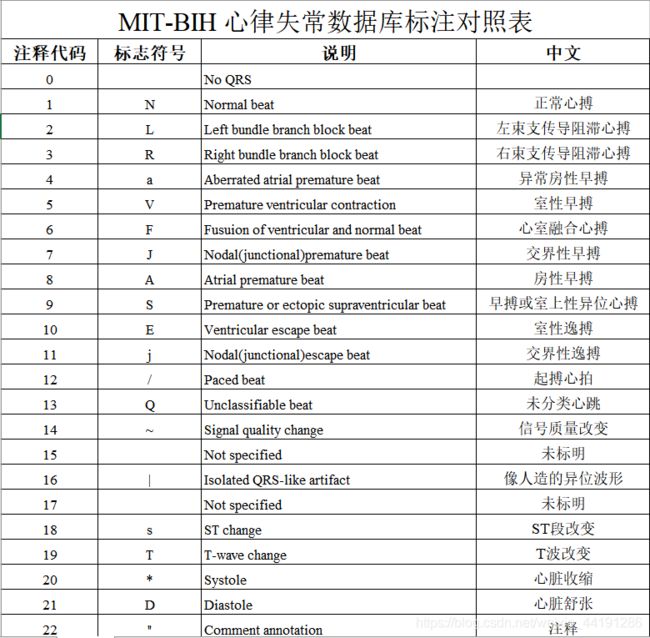
在ann的标记中,一共有44种标记类型(如表所示),其中是R波的标记位置只有19种,所以如果要进行R波位置的读取还需要将不需要的标记给去掉

def delete_non_beat(symbol, sample, **kw):
"""删除非心拍的标注符号"""
AAMI_MIT_MAP = {'N': 'Nfe/jnBLR',#将19类信号分为五大类
'S': 'SAJa',
'V': 'VEr',
'F': 'F',
'Q': 'Q?'}
MIT2AAMI = {c: k for k in AAMI_MIT_MAP.keys() for c in AAMI_MIT_MAP[k]}
mit_beat_codes = list(MIT2AAMI.keys())
symbol = np.array(symbol)#symbol对应标签,sample为R峰所在位置,sig为R峰值
isin = np.isin(symbol, mit_beat_codes)
symbol = symbol[isin]#去除19类之外的非心拍标注信号
symbol = np.array([MIT2AAMI[x] for x in symbol])#将19类信号划分为五类
return symbol, np.copy(sample[isin])#sample对应R峰采样点位置,利用R峰值判断AAMI五种类型
持续更新ing
wfdb.processing.normalize_bound(sig, lb=0, ub=1)
Normalize a signal between the lower and upper bound
sig : numpy array
Original signal to be normalized
lb : int, or float
Lower bound
ub : int, or float
Upper bound
x_normalized : numpy array
Normalized signal
wfdb.processing.get_filter_gain(b, a, f_gain, fs)
Given filter coefficients, return the gain at a particular frequency.
b : list
List of linear filter b coefficients
a : list
List of linear filter a coefficients
f_gain : int or float, optional
The frequency at which to calculate the gain
fs : int or float, optional
The sampling frequency of the system
参考文档:
1: Demo Scripts for the wfdb-python package
2: wfdb 2.2.1 documentation