医学图像处理(一)使用U-Net进行MRI的肝脏分割
医学图像处理(一)使用U-Net进行MRI的肝脏分割
- 数据集搜集
- 训练集和测试集划分
-
- 问题一: 测试集不包括GroundTruth
- 问题二:T1和T2的数据能一起训练吗?
- 问题三: T1的图像在文件夹中分成了InPhase和OutPhase,这个有什么区别?
- 分离出肝脏
- 将dicom文件转换为png格式
- 数据增强
- U-net网络搭建
- 自定义Dataset
- Main.py
- 实验结果
- 总结
图像分割小白一枚,之前接到一个MRI图像肝脏分割的任务。对于医学图像分割,第一想到的就是Unet. Unet在2015的MICCAI上首次提出,在医学数据集上效果很好,因而成为大多做医疗影像语义分割任务的baseline. 关于Unet网络结构,网上有许多详实的资料对其解析,我也不班门弄斧,只是简单记录一下我的学习和实践经历。
Github地址: https://github.com/BUAAXZzz/Unet_liver_seg
说明文档和requirements.txt还在完善中…
数据集搜集
第一步当然是寻找数据集。在许多图像分类比赛中有免费开源的数据集,推荐一个网站:https://grand-challenge.org/challenges/. 它归总了至今为止大部分的挑战赛及其数据集,只不过有的数据可能下不动,需要科学上网.
这是港中文的窦琪学姐在线上报告上总结的各类开源数据集.

CHAOS数据集的附带文献也列举了近些年各类Challenges所公开的数据集(截止2019年).

CHAOS数据集拥有腹腔的MRI图像,它包括脾脏、肝脏、左肾和右肾等四个器官的MRI图像和Ground Truth.
CHAOS数据集下载地址:https://chaos.grand-challenge.org/Download/
该数据集下载很慢,需要科学上网。我是在某宝上花了2块钱购买的下载服务。
我这里也分享一个百度网盘的地址:
链接:https://pan.baidu.com/s/1OSbgU_Bxp85dDv-ElSzrVw
提取码:x2z3
训练集和测试集划分
问题一: 测试集不包括GroundTruth
下载下来后,有CT/MR两种数据,都是dcm格式,每一张就是一个slice. 对于核磁共振图像,一共有40个病例,训练集和测试集各分了20例。比较坑的是,测试集并没有给出GroundTruth的图像,可能是比赛需要吧. 因而我们只能将使用一半的病例,并将其重新划分成训练和测试集。
我使用了16个病例做训练集,4个病例做测试集.
问题二:T1和T2的数据能一起训练吗?
据我的实验结果来看,仅仅用Unet分割是不能一起训练的,最终的结果会导致严重的过拟合;T1,T2图像可能在模态上还是有比较大的区别.
我对医学不懂,查阅资料:
- T1加权成像(T1WI)是指突出组织T纵向弛豫差别。t1越短,指信号越强,t1越长,指信号越弱,t1一般用于观察解剖。
- T2加权成像(T2WI)是指突出组织T2横向弛豫差别。t2越短,是指信号越弱,t2越长,则信号越强,一般t2有利于观察病变,对出血较敏感。
最终我选择使用T1的图像训练.
问题三: T1的图像在文件夹中分成了InPhase和OutPhase,这个有什么区别?
InPhase和Outphase应该是T1模态图像的相位相反的两种呈现,我只选用了InPhase进行了训练,因为它跟GroundTruth的命名是一样的,处理起来也方便一点.
下面是分离数据集的代码:
"""
@ Date: 2020/6/29
@ Author: Xiao Zhuo
@ Brief: Split CHAOS DataSet into my directory
@ Filename: split_dataset_1.py
"""
# -*- coding: utf-8 -*-
import os
import shutil
import random
dst_TrainData = "./data/train/Data"
dst_TrainGround = "./data/train/Ground"
dst_TestData = "./data/val/Data"
dst_TestGround = "./data/val/Ground"
def collect_T1_name(patient_dir):
ground_paths = list()
inphase_paths = list()
t1_datadir = os.path.join(patient_dir, "T1DUAL")
ground_dir = os.path.join(t1_datadir, "Ground")
ground_names = os.listdir(ground_dir)
nums_ground = len(ground_names)
# 拼接Ground文件夹的文件,存入到ground_paths列表中
for i in range(nums_ground):
ground_paths.append(os.path.join(ground_dir, ground_names[i]))
inphase_dir = os.path.join(t1_datadir, "DICOM_anon", "InPhase")
inphase_names = os.listdir(inphase_dir)
nums_inphase = len(inphase_names)
# 拼接inphase文件夹的文件,存入到inphase_paths列表中
for i in range(nums_inphase):
inphase_paths.append(os.path.join(inphase_dir, inphase_names[i]))
return ground_paths, inphase_paths
if __name__ == '__main__':
dataset_dir = os.path.join("CHAOS_Train_Sets", "Train_Sets", "MR")
train_pct = 0.8
test_pct = 0.2
for root, dirs, files in os.walk(dataset_dir):
random.shuffle(dirs)
dir_count = len(dirs)
train_point = int(dir_count * train_pct)
i = 0
for sub_dir in dirs: # sub_dir代表病人编号
if i < train_point:
patient_dir = os.path.join(root, sub_dir)
ground_paths, inphase_paths = collect_T1_name(patient_dir)
for num in range(len(ground_paths)):
dst_groundpath = os.path.join(dst_TrainGround, "T1_Patient%s_No%d.png" % (sub_dir, num))
shutil.copy(ground_paths[num], dst_groundpath)
## 下面待修改
for num in range(len(inphase_paths)):
dst_inphasepath = os.path.join(dst_TrainData, "T1_Patient%s_No%d.dcm" % (sub_dir, num))
shutil.copy(inphase_paths[num], dst_inphasepath)
i += 1
else:
patient_dir = os.path.join(root, sub_dir)
ground_paths, inphase_paths = collect_T1_name(patient_dir)
for num in range(len(ground_paths)):
dst_groundpath = os.path.join(dst_TestGround, "T1_Patient%s_No%d.png" % (sub_dir, num))
shutil.copy(ground_paths[num], dst_groundpath)
for num in range(len(inphase_paths)):
dst_inphasepath = os.path.join(dst_TestData, "T1_Patient%s_No%d.dcm" % (sub_dir, num))
shutil.copy(inphase_paths[num], dst_inphasepath)
i += 1
该段程序的作用就是将训练集中T1/InPhase 的20个病例划分成16个训练集,4个测试集,并重新存储到自定义的文件夹下. 对于图像文件也进行了命名规范,对第i个病人的第j张slice,命名规则为T1_Patienti_Noj.dcm
分离出肝脏
GroundTruth的图像是多器官的,根据灰度范围进行判断。

从自带的config文件中可以查看灰度范围:

因为GroundTruth是png格式,我们使用OpenCV做一下简单的阈值处理就可以提取肝脏部分了.
"""
@ Date: 2020/6/29
@ Author: Xiao Zhuo
@ Brief: Extract liver part from GroundTruth and set white color
@ Filename: extract_only_liver_2.py
"""
# -*- coding: utf-8 -*-
import os
import cv2
def makedir(dir):
if not os.path.exists(dir):
os.mkdir(dir)
def extract_liver(dataset_dir):
src_names = os.listdir(dataset_dir)
if src_names[0] == 'Liver':
src_names.remove('Liver')
src_count = len(src_names)
dst_dir = os.path.join(dataset_dir, "Liver")
makedir(dst_dir)
for num in range(src_count):
src_path = os.path.join(dataset_dir, src_names[num])
src = cv2.imread(src_path) # OpenCV读进来要指定是灰度图像,不然默认三通道。这里之前忘记指定了
# flag = 0
flag = 1
for i in range(src.shape[0]):
for j in range(src.shape[1]):
for k in range(src.shape[2]):
if 55 <= src.item(i, j, k) <= 70:
flag = 1 # 表示有肝脏
src.itemset((i, j, k), 255)
else:
src.itemset((i, j, k), 0)
if flag == 1:
dst_path = os.path.join(dst_dir, src_names[num])
cv2.imwrite(dst_path, src)
if __name__ == '__main__':
train_dir = os.path.join("data", "train", "Ground")
test_dir = os.path.join("data", "val", "Ground")
extract_liver(train_dir)
extract_liver(test_dir)
将dicom文件转换为png格式
这一步实际上也可以不做,原因是dicom中的图像数据原本是16位的,若是转换成8位的png格式可能会导致数据精度丢失。使用SimpleITK直接读取Array送入U-net其实就可以运行了.
但我要多此一举的原因是,我想做数据增强. 但是现有的数据增强工具好像不能处理Array或者numpy等格式的数据,自己又没有那个水平重新写一个数据增强的API。没办法,就转换成png简单处理吧.
"""
# @file name : conver2png.py
# @author : Peter
# @date : 2020-07-01
# @brief : 将dicom格式转换成png格式
"""
import pydicom
import os
import matplotlib.pyplot as plt
from skimage import img_as_float
path_1 = "./data/val/Data"
path_2 = "./data/train/Data"
def dicom_2png(orifile, savefile, width, height):
_currFile = orifile
dcm = pydicom.dcmread(orifile)
# fileName = os.path.basename(file)
imageX = dcm.pixel_array
temp = imageX.copy()
picMax = imageX.max()
vmin = imageX.min()
vmax = temp[temp < picMax].max()
# print("vmin : ", vmin)
# print("vmax : ", vmax)
imageX[imageX > vmax] = 0
imageX[imageX < vmin] = 0
# result = exposure.is_low_contrast(imageX)
# # print(result)
image = img_as_float(imageX)
plt.cla()
plt.figure('adjust_gamma', figsize=(width/100, height/100))
plt.subplots_adjust(top=1, bottom=0, left=0, right=1, hspace=0, wspace=0)
plt.imshow(image, 'gray')
plt.axis('off')
plt.savefig(savefile)
if __name__ == '__main__':
names = os.listdir(path_1)
for i in range(len(names)):
dicom_path = os.path.join(path_1, names[i])
png_name = os.path.splitext(names[i])[0]
dst_path = os.path.join('./data/val/Data_8bit', (png_name + '.png'))
dicom_2png(dicom_path, dst_path, 256, 256)
names = os.listdir(path_2)
for i in range(len(names)):
dicom_path = os.path.join(path_2, names[i])
png_name = os.path.splitext(names[i])[0]
dst_path = os.path.join('./data/train/Data_8bit', (png_name + '.png'))
dicom_2png(dicom_path, dst_path, 256, 256)
转换后一目了然,不需要再用MicroDicom去查看
数据增强
我使用Augmentor工具.
# 导入数据增强工具
import Augmentor
# 确定原始图像存储路径以及掩码文件存储路径
p = Augmentor.Pipeline("./data/train/Data")
p.ground_truth("./data/train/Ground")
# 图像旋转: 按照概率0.8执行,最大左旋角度10,最大右旋角度10
p.rotate(probability=0.8, max_left_rotation=10, max_right_rotation=10)
# 图像左右互换: 按照概率0.5执行
p.flip_left_right(probability=0.5)
# 图像放大缩小: 按照概率0.8执行,面积为原始图0.85倍
p.zoom_random(probability=0.3, percentage_area=0.85)
# 最终扩充的数据样本数
p.sample(400)
当然,增强的图片还可以重新命个名,按照序号来:
import os
Data_path = "./data/train/Data_aug"
Ground_path = "./data/train/Ground_aug"
data_names = os.listdir(Data_path)
ground_names = os.listdir(Ground_path)
for i in range(len(data_names)):
used_name = os.path.join(Data_path, data_names[i])
new_name = os.path.join(Data_path, "Aug_No_%d.png" % i)
os.rename(used_name, new_name)
for i in range(len(ground_names)):
used_name = os.path.join(Ground_path, ground_names[i])
new_name = os.path.join(Ground_path, "Aug_No_%d.png" % i)
os.rename(used_name, new_name)
网络搭建和训练部分,我使用的是Python3.7 + Pytorch 1.4.0.
U-net网络搭建
就是经典的网络结构,不过我加了尝试加了几个Dropout层.
"""
@ filename: unet.py
"""
import torch
from torch import nn
class DoubleConv(nn.Module):
def __init__(self, in_ch, out_ch):
super(DoubleConv, self).__init__()
self.conv = nn.Sequential(
nn.Conv2d(in_ch, out_ch, 3, padding=1),
nn.BatchNorm2d(out_ch),
nn.ReLU(inplace=True),
nn.Conv2d(out_ch, out_ch, 3, padding=1),
nn.BatchNorm2d(out_ch),
nn.ReLU(inplace=True)
)
def forward(self, input):
return self.conv(input)
class Unet(nn.Module):
def __init__(self, in_ch, out_ch):
super(Unet, self).__init__()
self.conv1 = DoubleConv(in_ch, 64)
self.pool1 = nn.MaxPool2d(2)
self.conv2 = DoubleConv(64, 128)
self.pool2 = nn.MaxPool2d(2)
self.conv3 = DoubleConv(128, 256)
self.pool3 = nn.MaxPool2d(2)
self.conv4 = DoubleConv(256, 512)
self.pool4 = nn.MaxPool2d(2)
self.conv5 = DoubleConv(512, 1024)
self.up6 = nn.ConvTranspose2d(1024, 512, 2, stride=2)
self.conv6 = DoubleConv(1024, 512)
self.up7 = nn.ConvTranspose2d(512, 256, 2, stride=2)
self.conv7 = DoubleConv(512, 256)
self.up8 = nn.ConvTranspose2d(256, 128, 2, stride=2)
self.conv8 = DoubleConv(256, 128)
self.up9 = nn.ConvTranspose2d(128, 64, 2, stride=2)
self.conv9 = DoubleConv(128, 64)
self.conv10 = nn.Conv2d(64, out_ch, 1)
self.dropout = nn.Dropout(p=0.2)
def forward(self, x):
c1 = self.conv1(x)
p1 = self.pool1(c1)
p1 = self.dropout(p1)
c2 = self.conv2(p1)
p2 = self.pool2(c2)
p2 = self.dropout(p2)
c3 = self.conv3(p2)
p3 = self.pool3(c3)
p3 = self.dropout(p3)
c4 = self.conv4(p3)
p4 = self.pool4(c4)
p4 = self.dropout(p4)
c5 = self.conv5(p4)
up_6 = self.up6(c5)
merge6 = torch.cat([up_6, c4], dim=1)
merge6 = self.dropout(merge6)
c6 = self.conv6(merge6)
up_7 = self.up7(c6)
merge7 = torch.cat([up_7, c3], dim=1)
merge7 = self.dropout(merge7)
c7 = self.conv7(merge7)
up_8 = self.up8(c7)
merge8 = torch.cat([up_8, c2], dim=1)
merge8 = self.dropout(merge8)
c8 = self.conv8(merge8)
up_9 = self.up9(c8)
merge9 = torch.cat([up_9, c1], dim=1)
merge9 = self.dropout(merge9)
c9 = self.conv9(merge9)
c10 = self.conv10(c9)
# out = nn.Sigmoid()(c10)
return c10
自定义Dataset
make_dataset方法获取原始图像和分割掩膜的图像路径名,LiverDateset类继承torch的数据集类,通过make_dataset的路径名利用PIL Image库读取文件,并进行transforms变换成归一化的Tensor数据.
"""
@ filename: dataset.py
@ author: Peter Xiao
@ Date: 2020/5/1
@ Brief: 自定义肝脏数据集
"""
from torch.utils.data import Dataset
import PIL.Image as Image
import os
def make_dataset(root):
# root = "./data/train"
imgs = []
ori_path = os.path.join(root, "Data")
ground_path = os.path.join(root, "Ground")
names = os.listdir(ori_path)
n = len(names)
for i in range(n):
img = os.path.join(ori_path, names[i])
mask = os.path.join(ground_path, names[i])
imgs.append((img, mask))
return imgs
class LiverDataset(Dataset):
def __init__(self, root, transform=None, target_transform=None):
imgs = make_dataset(root)
self.imgs = imgs
self.transform = transform
self.target_transform = target_transform
def __getitem__(self, index):
x_path, y_path = self.imgs[index]
img_x = Image.open(x_path).convert('L')
img_y = Image.open(y_path).convert('L')
if self.transform is not None:
img_x = self.transform(img_x)
if self.target_transform is not None:
img_y = self.target_transform(img_y)
return img_x, img_y
def __len__(self):
return len(self.imgs)
Main.py
Main文件主要有三个功能,训练、预测(包括生成可视化图像)和计算Dice系数. 主程序利用了argparse模块作命令行,可以自行修改.
这里提醒一点:我训练时使用的GPU是GTX1650,显存4G. batch_size设在4刚刚好,调大了会爆显存,无法训练. 在实验室的2080Ti上用16的BT训练,占用显存为9.1G,可以根据这个比例结合自己的GPU调整Batch_size.
"""
@ filename: main.py
@ author: Peter Xiao
@ date: 2020/5/1
@ brief: MR肝脏分割,训练、测试和计算Dice系数
"""
import torch
import argparse
import cv2
import os
import numpy as np
import matplotlib.pyplot as plt
from torch.utils.data import DataLoader
from torch import nn, optim
from torchvision.transforms import transforms
from unet import Unet
from denseunet import DenseUNet_65, DenseUNet_167
from dataset import LiverDataset
from tools.common_tools import transform_invert
val_interval = 1
# 是否使用cuda
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
x_transforms = transforms.Compose([
transforms.ToTensor(),
])
# mask只需要转换为tensor
y_transforms = transforms.ToTensor()
train_curve = list()
valid_curve = list()
def train_model(model, criterion, optimizer, dataload, num_epochs=80):
model_path = "./model/Aug/weights_20.pth"
if os.path.exists(model_path):
model.load_state_dict(torch.load(model_path, map_location=device))
start_epoch = 20
print('加载成功!')
else:
start_epoch = 0
print('无保存模型,将从头开始训练!')
for epoch in range(start_epoch+1, num_epochs):
print('Epoch {}/{}'.format(epoch, num_epochs))
print('-' * 10)
dt_size = len(dataload.dataset)
epoch_loss = 0
step = 0
for x, y in dataload:
step += 1
inputs = x.to(device)
labels = y.to(device)
# zero the parameter gradients
optimizer.zero_grad()
# forward
outputs = model(inputs)
loss = criterion(outputs, labels)
loss.backward()
optimizer.step()
epoch_loss += loss.item()
train_curve.append(loss.item())
print("%d/%d,train_loss:%0.3f" % (step, (dt_size - 1) // dataload.batch_size + 1, loss.item()))
print("epoch %d loss:%0.3f" % (epoch, epoch_loss/step))
if (epoch + 1) % 20 == 0:
torch.save(model.state_dict(), './model/Aug/weights_%d.pth' % (epoch + 1))
# Validate the model
valid_dataset = LiverDataset("data/val", transform=x_transforms, target_transform=y_transforms)
valid_loader = DataLoader(valid_dataset, batch_size=4, shuffle=True)
if (epoch + 2) % val_interval == 0:
loss_val = 0.
model.eval()
with torch.no_grad():
step_val = 0
for x, y in valid_loader:
step_val += 1
inputs = x.to(device)
labels = y.to(device)
outputs = model(inputs)
loss = criterion(outputs, labels)
loss_val += loss.item()
valid_curve.append(loss_val)
print("epoch %d valid_loss:%0.3f" % (epoch, loss_val / step_val))
train_x = range(len(train_curve))
train_y = train_curve
train_iters = len(dataload)
valid_x = np.arange(1, len(
valid_curve) + 1) * train_iters * val_interval # 由于valid中记录的是EpochLoss,需要对记录点进行转换到iterations
valid_y = valid_curve
plt.plot(train_x, train_y, label='Train')
plt.plot(valid_x, valid_y, label='Valid')
plt.legend(loc='upper right')
plt.ylabel('loss value')
plt.xlabel('Iteration')
plt.show()
return model
#训练模型
def train(args):
model = Unet(1, 1).to(device)
# model = DenseUNet_65(1, 1).to(device)
batch_size = args.batch_size
criterion = nn.BCEWithLogitsLoss()
optimizer = optim.Adam(model.parameters())
liver_dataset = LiverDataset("./data/train", transform=x_transforms, target_transform=y_transforms)
dataloaders = DataLoader(liver_dataset, batch_size=batch_size, shuffle=True, num_workers=4)
train_model(model, criterion, optimizer, dataloaders)
#显示模型的输出结果
def test(args):
model = Unet(1, 1)
model.load_state_dict(torch.load(args.ckpt, map_location='cuda'))
liver_dataset = LiverDataset("data/val", transform=x_transforms, target_transform=y_transforms)
dataloaders = DataLoader(liver_dataset, batch_size=1)
save_root = "E:\\MyDocuments\\TorchLearing\\u_net_liver_chaos_8bit\\data\\predict\\test"
model.eval()
plt.ion()
index = 0
with torch.no_grad():
for x, ground in dataloaders:
y = model(x)
x = torch.squeeze(x)
x = x.unsqueeze(0)
ground = torch.squeeze(ground)
ground = ground.unsqueeze(0)
img_ground = transform_invert(ground, y_transforms)
img_x = transform_invert(x, x_transforms)
img_y = torch.squeeze(y).numpy()
# cv2.imshow('img', img_y)
src_path = os.path.join(save_root, "predict_%d_s.png" % index)
save_path = os.path.join(save_root, "predict_%d_o.png" % index)
ground_path = os.path.join(save_root, "predict_%d_g.png" % index)
img_ground.save(ground_path)
img_x.save(src_path)
cv2.imwrite(save_path, img_y * 255)
index = index + 1
# plt.imshow(img_y)
# plt.pause(0.5)
# plt.show()
# 计算Dice系数
def dice_calc(args):
root = "E:\\MyDocuments\\TorchLearing\\u_net_liver_chaos_8bit\\data\\predict\\aug+drop_8bit\\epoch80"
nums = len(os.listdir(root)) // 3
dice = list()
dice_mean = 0
for i in range(nums):
ground_path = os.path.join(root, "predict_%d_g.png" % i)
predict_path = os.path.join(root, "predict_%d_o.png" % i)
img_ground = cv2.imread(ground_path)
img_predict = cv2.imread(predict_path)
intersec = 0
x = 0
y = 0
for w in range(256):
for h in range(256):
intersec += img_ground.item(w, h, 1) * img_predict.item(w, h, 1) / (255 * 255)
x += img_ground.item(w, h, 1) / 255
y += img_predict.item(w, h, 1) / 255
if x + y == 0:
current_dice = 1
else:
current_dice = round(2 * intersec / (x + y), 3)
dice_mean += current_dice
dice.append(current_dice)
dice_mean /= len(dice)
print(dice)
print(round(dice_mean, 3))
if __name__ == '__main__':
#参数解析
parse = argparse.ArgumentParser()
parse.add_argument("--action", type=str, help="train, test or dice", default="test")
parse.add_argument("--batch_size", type=int, default=4)
parse.add_argument("--ckpt", type=str, help="the path of model weight file", default="./model/Aug/weights_80.pth")
# parse.add_argument("--ckpt", type=str, help="the path of model weight file")
args = parse.parse_args()
if args.action == "train":
train(args)
elif args.action == "test":
test(args)
elif args.action == "dice":
dice_calc(args)
实验结果
训练速度还是很快的,GTX1650在Batch_size为4的情况下训练20个epoch的时间在20分组以内. 20个Epoch的结果如下:横向的连续三张图分别为GroundTruth,网络预测图及原图。看起来还是不错的.
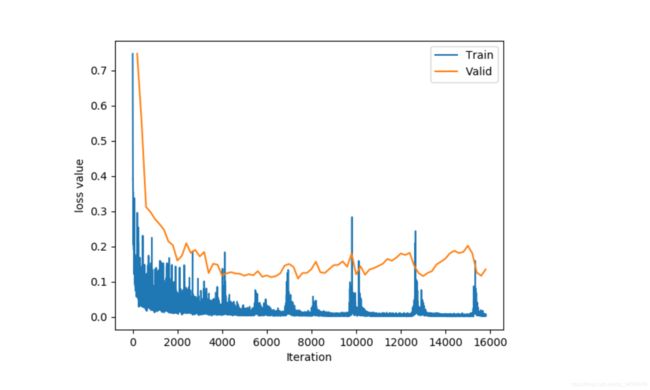
另外我使用了原始的16位数据训练,发现Dice并不是很好,不知道是什么原因。同时我还对训练时间、是否数据增强、是否添加dropout等进行了对比实验,Dice箱线图如下:

Dice最佳为0.89,离CHAOS文献中2D-Unet的最好结果91%还有一些距离,不过我只使用了16个病例,而且Unet基本没有改动,这个结果还是比较正常了.
总结
实验主要是在原始数据的处理上耗费了比较多的功夫,对os/shutil库,文件的复制移动以及图像格式的转换需要比较熟练。网络是现成的,因此在训练这块我没有花太多功夫。这也正印证了数据的重要性。有时候良好的数据+简单的网络训练出来往往比较差的数据+最新的网络要好很多。
