写在前面
本文章参考了多个国内外教程,在这里就不列出来了。这么写的原因还是想告诉大家:本文章并非原创,感谢各位前辈的分享。
加载需要的数据包
本文章用到的数据包中,“tidyverse”,“skimr”,“FactoMineR”,“factoextra”,“pheatmap”是必须的。但是为了更好的查看数据,所以又加入了“GGally”,“patchwork”,"ggstatsplot和"“ggpubr”。后几个包并不是必须的,但会让你的数据可视化更方便。各种包按自己的好恶来,例如有人就极度讨厌“ggpubr”。最后“pheatmap”虽然已经被作者放弃了,但是我觉得他搞的“ComplexHeatmap”实在是有点走火入魔……可能他也意识到这个问题,现在给了个CompleHeatmap::pheatmap的选项(还能说什么,绝了)。什么你问knitr干嘛的?你猜……
knitr::opts_chunk$set(echo = TRUE, warning = FALSE)
library('tidyverse')
library('ggpubr')
library('patchwork')
library('skimr')
library('GGally')
library('lme4')
library("FactoMineR")
library("factoextra")
library('ComplexHeatmap')
library('ggstatsplot')
library('agricolae')
library('car')
library('vip')
library('onewaytests')
library('jmv')
读取数据
这里rm命令清空存在环境中的所有变量,避免先前环境中的变量对接下来的操作带来影响。
rm(list = ls())
为了方便大家测试,这里使用了R语言自带数据集iris,如果你想用mtcars或者别的请随意。比如想看汽车各种特性对油耗的影响就可以用mtcars。skim可以很华丽的展示你的数据结构。当然,没啥别的用途了。
head(iris)
data <- iris
skim(data %>% group_by(Species))
| skim_variable | Species | n_missing | complete_rate | mean | sd | p0 | p25 | p50 | p75 | p100 | hist |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sepal.Length | setosa | 0 | 1 | 5.01 | 0.35 | 4.3 | 4.80 | 5.00 | 5.20 | 5.8 | ▃▃▇▅▁ |
| Sepal.Length | versicolor | 0 | 1 | 5.94 | 0.52 | 4.9 | 5.60 | 5.90 | 6.30 | 7.0 | ▂▇▆▃▃ |
| Sepal.Length | virginica | 0 | 1 | 6.59 | 0.64 | 4.9 | 6.23 | 6.50 | 6.90 | 7.9 | ▁▃▇▃▂ |
| Sepal.Width | setosa | 0 | 1 | 3.43 | 0.38 | 2.3 | 3.20 | 3.40 | 3.68 | 4.4 | ▁▃▇▅▂ |
| Sepal.Width | versicolor | 0 | 1 | 2.77 | 0.31 | 2.0 | 2.52 | 2.80 | 3.00 | 3.4 | ▁▅▆▇▂ |
| Sepal.Width | virginica | 0 | 1 | 2.97 | 0.32 | 2.2 | 2.80 | 3.00 | 3.18 | 3.8 | ▂▆▇▅▁ |
| Petal.Length | setosa | 0 | 1 | 1.46 | 0.17 | 1.0 | 1.40 | 1.50 | 1.58 | 1.9 | ▁▃▇▃▁ |
| Petal.Length | versicolor | 0 | 1 | 4.26 | 0.47 | 3.0 | 4.00 | 4.35 | 4.60 | 5.1 | ▂▂▇▇▆ |
| Petal.Length | virginica | 0 | 1 | 5.55 | 0.55 | 4.5 | 5.10 | 5.55 | 5.88 | 6.9 | ▃▇▇▃▂ |
| Petal.Width | setosa | 0 | 1 | 0.25 | 0.11 | 0.1 | 0.20 | 0.20 | 0.30 | 0.6 | ▇▂▂▁▁ |
| Petal.Width | versicolor | 0 | 1 | 1.33 | 0.20 | 1.0 | 1.20 | 1.30 | 1.50 | 1.8 | ▅▇▃▆▁ |
| Petal.Width | virginica | 0 | 1 | 2.03 | 0.27 | 1.4 | 1.80 | 2.00 | 2.30 | 2.5 | ▂▇▆▅▇ |
随时随地的可视化
这是我认为R语言跟spss和其他软件比最大的优势了。是的,在Rstudio中你可以随时随地的可视化,无限制的切片数据可视化。相比于单纯的统计分析,我认为视觉往往来的更准确更直接。话不多说让我们开始吧。
首先,我们在R语言里进行一些传统的方法。我们进行一个切片求平均值。这里我们使用了tidyverse套件(dplyr)。其中的 %>% 是通道符号,他的含义是将前面的参数传入到下一个命令中作为第一个参数(可以用.代替)。统计各个组的平均值和标准偏差,过去大家都在用spss得到这些,统计分析,最后获得结果,大家都满意了,在R里你同样可以做到。
data_summary <- data %>% group_by(Species) %>% summarise_each(funs(mean,sd),
Sepal.Length, Sepal.Width,
Petal.Length, Petal.Width)
data_summary
## # A tibble: 3 x 9
## Species Sepal.Length_me~ Sepal.Width_mean Petal.Length_me~ Petal.Width_mean
##
## 1 setosa 5.01 3.43 1.46 0.246
## 2 versic~ 5.94 2.77 4.26 1.33
## 3 virgin~ 6.59 2.97 5.55 2.03
## # ... with 4 more variables: Sepal.Length_sd , Sepal.Width_sd ,
## # Petal.Length_sd , Petal.Width_sd
当然你也可以这样……
data_res <- data %>% pivot_longer(col = -Species, names_to = 'Name', values_to = 'Value') %>%
group_by(Species,Name) %>%
summarise(mean = mean(Value), sd = mean(Value))
data_res
## # A tibble: 12 x 4
## # Groups: Species [3]
## Species Name mean sd
##
## 1 setosa Petal.Length 1.46 1.46
## 2 setosa Petal.Width 0.246 0.246
## 3 setosa Sepal.Length 5.01 5.01
## 4 setosa Sepal.Width 3.43 3.43
## 5 versicolor Petal.Length 4.26 4.26
## 6 versicolor Petal.Width 1.33 1.33
## 7 versicolor Sepal.Length 5.94 5.94
## 8 versicolor Sepal.Width 2.77 2.77
## 9 virginica Petal.Length 5.55 5.55
## 10 virginica Petal.Width 2.03 2.03
## 11 virginica Sepal.Length 6.59 6.59
## 12 virginica Sepal.Width 2.97 2.97
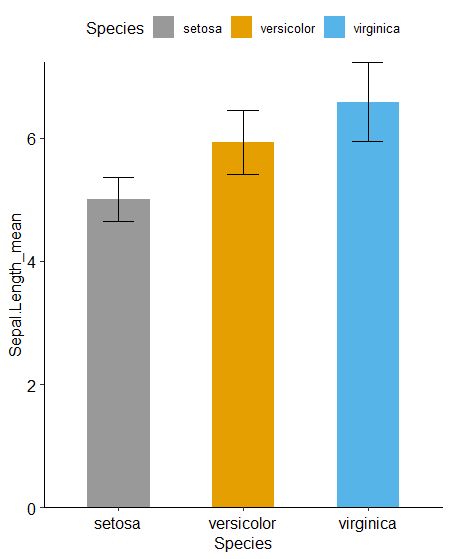
有了平均值和标准差,我们自然能画出第一个柱状图。
cbPalette <- c("#999999", "#E69F00", "#56B4E9", "#009E73", "#F0E442", "#0072B2", "#D55E00", "#CC79A7")
p1 <- ggplot(data_summary,aes(x = Species, y = Sepal.Length_mean, fill = Species)) +
geom_bar(stat = 'identity', position = 'dodge', width = 0.5) +
geom_errorbar(aes(ymin = Sepal.Length_mean - Sepal.Length_sd,
ymax = Sepal.Length_mean + Sepal.Length_sd), width = 0.25) +
scale_fill_manual(values = cbPalette) +
scale_y_continuous(expand = c(0, 0)) +
theme_pubr()
p1
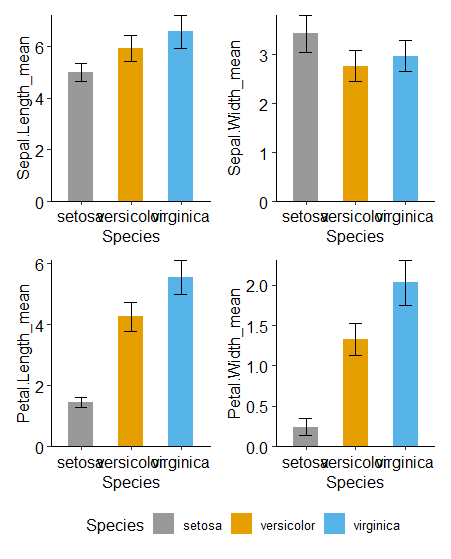
重复四次,你能获得四个柱状图
compare_group <- list(c('setosa','versicolor'),c('versicolor','virginica'),c('setosa','virginica'))
p1 <- ggplot(data_summary,aes(x = Species, y = Sepal.Length_mean, fill = Species)) +
geom_bar(stat = 'identity', position = 'dodge', width = 0.5) +
geom_errorbar(aes(ymin = Sepal.Length_mean - Sepal.Length_sd,
ymax = Sepal.Length_mean + Sepal.Length_sd), width = 0.25) +
scale_fill_manual(values = cbPalette) +
scale_y_continuous(expand = c(0, 0)) +
theme_pubr()
p2 <- ggplot(data_summary,aes(x = Species, y = Sepal.Width_mean, fill = Species)) +
geom_bar(stat = 'identity', position = 'dodge', width = 0.5) +
geom_errorbar(aes(ymin = Sepal.Width_mean - Sepal.Width_sd,
ymax = Sepal.Width_mean + Sepal.Width_sd), width = 0.25) +
scale_fill_manual(values = cbPalette) +
scale_y_continuous(expand = c(0, 0)) +
theme_pubr()
p3 <- ggplot(data_summary,aes(x = Species, y = Petal.Length_mean, fill = Species)) +
geom_bar(stat = 'identity', position = 'dodge', width = 0.5) +
geom_errorbar(aes(ymin = Petal.Length_mean - Petal.Length_sd,
ymax = Petal.Length_mean + Petal.Length_sd), width = 0.25) +
scale_fill_manual(values = cbPalette) +
scale_y_continuous(expand = c(0, 0)) +
theme_pubr()
p4 <- ggplot(data_summary,aes(x = Species, y = Petal.Width_mean, fill = Species)) +
geom_bar(stat = 'identity', position = 'dodge', width = 0.5) +
geom_errorbar(aes(ymin = Petal.Width_mean - Petal.Width_sd,
ymax = Petal.Width_mean + Petal.Width_sd), width = 0.25) +
scale_fill_manual(values = cbPalette) +
scale_y_continuous(expand = c(0, 0)) +
theme_pubr()
p1+p2+p3+p4+plot_layout(guides = 'collect')&theme(legend.position = 'bottom')
然后你可以做统计分析。别忘了检查各个品种的变量是否满足正态分布,会发现正态性检验还好。除了setosa的petal.width不是很满足。
data %>% group_by(Species) %>%
summarise(statistic = shapiro.test(Sepal.Length)$statistic,
p.value = shapiro.test(Sepal.Length)$p.value)
data %>% group_by(Species) %>%
summarise(statistic = shapiro.test(Sepal.Width)$statistic,
p.value = shapiro.test(Sepal.Width)$p.value)
data %>% group_by(Species) %>%
summarise(statistic = shapiro.test(Petal.Length)$statistic,
p.value = shapiro.test(Petal.Length)$p.value)
data %>% group_by(Species) %>%
summarise(statistic = shapiro.test(Petal.Width)$statistic,
p.value = shapiro.test(Petal.Width)$p.value)
## # A tibble: 3 x 3
## Species statistic p.value
##
## 1 setosa 0.978 0.460
## 2 versicolor 0.978 0.465
## 3 virginica 0.971 0.258
## # A tibble: 3 x 3
## Species statistic p.value
##
## 1 setosa 0.972 0.272
## 2 versicolor 0.974 0.338
## 3 virginica 0.967 0.181
## # A tibble: 3 x 3
## Species statistic p.value
##
## 1 setosa 0.955 0.0548
## 2 versicolor 0.966 0.158
## 3 virginica 0.962 0.110
## # A tibble: 3 x 3
## Species statistic p.value
##
## 1 setosa 0.800 0.000000866
## 2 versicolor 0.948 0.0273
## 3 virginica 0.960 0.0870
方差齐性检验。发现各组petal.length和petal.width方差不齐。
leveneTest(Sepal.Length ~ Species, data=data)
leveneTest(Sepal.Width ~ Species, data=data)
leveneTest(Petal.Length ~ Species, data=data)
leveneTest(Petal.Width ~ Species, data=data)
## Levene's Test for Homogeneity of Variance (center = median)
## Df F value Pr(>F)
## group 2 6.3527 0.002259 **
## 147
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
## Levene's Test for Homogeneity of Variance (center = median)
## Df F value Pr(>F)
## group 2 0.5902 0.5555
## 147
## Levene's Test for Homogeneity of Variance (center = median)
## Df F value Pr(>F)
## group 2 19.48 3.129e-08 ***
## 147
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
## Levene's Test for Homogeneity of Variance (center = median)
## Df F value Pr(>F)
## group 2 19.892 2.261e-08 ***
## 147
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
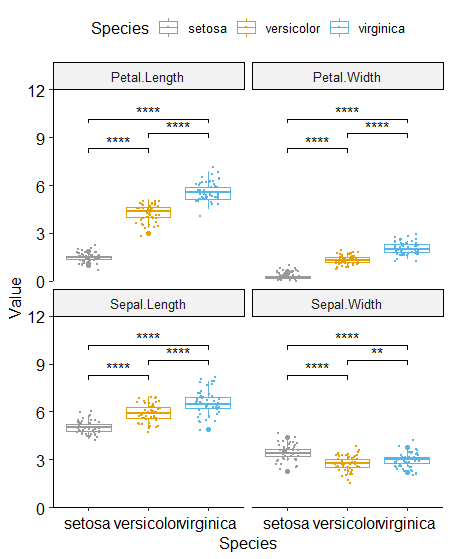
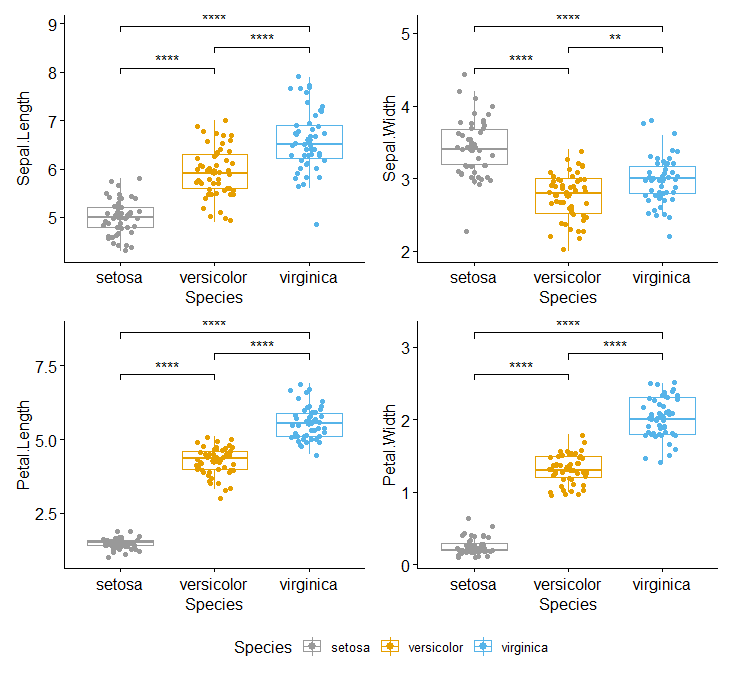
可以换成箱线图,再加上散点,标记好显著性。因为之前发现有点不满足正态和方差齐性,所以这里选择了非参数检验。
data %>% pivot_longer(col = -Species, names_to = 'Name', values_to = 'Value') %>%
ggplot(aes(x = Species, y = Value, color = Species)) +
geom_boxplot( ) +
geom_jitter(width = 0.2, height = 0.5, size = .1) +
scale_color_manual(values = cbPalette) +
scale_y_continuous(expand = c(0, 0),
limits = c(0, 12)) +
facet_wrap(~Name) +
stat_compare_means(comparisons = compare_group, label = 'p.signif',method = 'wilcox.test') + theme_pubr()
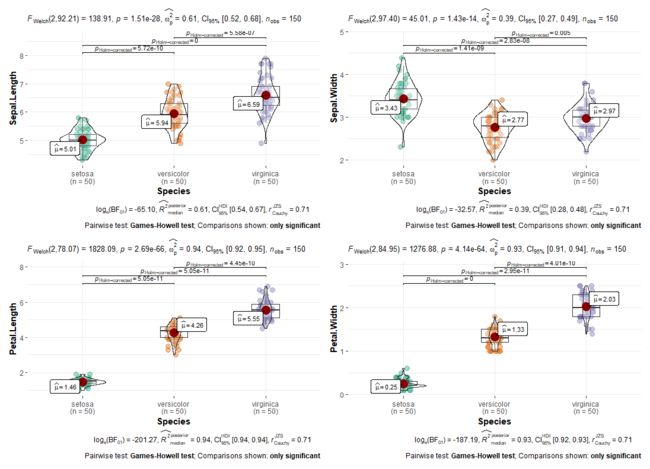
如果你觉得wilcox.test还不够稳健,你可以试试Welch's ANOVA检验和Games-Howell事后检验
anovaOneW(deps=Sepal.Length,group=Species,data=data,desc=T,
descPlot = T,norm=T,qq=T,eqv=T,phMeanDif = T,
phMethod= "gamesHowell",phTest = T,phFlag=T)
anovaOneW(deps=Sepal.Width,group=Species,data=data,desc=T,
descPlot = T,norm=T,qq=T,eqv=T,phMeanDif = T,
phMethod= "gamesHowell",phTest = T,phFlag=T)
anovaOneW(deps=Petal.Length,group=Species,data=data,desc=T,
descPlot = T,norm=T,qq=T,eqv=T,phMeanDif = T,
phMethod= "gamesHowell",phTest = T,phFlag=T)
anovaOneW(deps=Petal.Width,group=Species,data=data,desc=T,
descPlot = T,norm=T,qq=T,eqv=T,phMeanDif = T,
phMethod= "gamesHowell",phTest = T,phFlag=T)
篇幅原因这里只放一部分结果
##
## ONE-WAY ANOVA
##
## One-Way ANOVA (Welch's)
## -------------------------------------------------------------
## F df1 df2 p
## -------------------------------------------------------------
## Sepal.Length 138.9083 2 92.21115 < .0000001
## -------------------------------------------------------------
##
##
## Group Descriptives
## ---------------------------------------------------------------------------
## Species N Mean SD SE
## ---------------------------------------------------------------------------
## Sepal.Length setosa 50 5.006000 0.3524897 0.04984957
## versicolor 50 5.936000 0.5161711 0.07299762
## virginica 50 6.588000 0.6358796 0.08992695
## ---------------------------------------------------------------------------
##
##
## ASSUMPTION CHECKS
##
## Normality Test (Shapiro-Wilk)
## ------------------------------------------
## W p
## ------------------------------------------
## Sepal.Length 0.9878974 0.2188639
## ------------------------------------------
## Note. A low p-value suggests a
## violation of the assumption of
## normality
##
##
## Homogeneity of Variances Test (Levene's)
## -------------------------------------------------------
## F df1 df2 p
## -------------------------------------------------------
## Sepal.Length 7.381092 2 147 0.0008818
## -------------------------------------------------------
##
##
## POST HOC TESTS
##
## Games-Howell Post-Hoc Test – Sepal.Length
## --------------------------------------------------------------------------
## setosa versicolor virginica
## --------------------------------------------------------------------------
## setosa Mean difference — -0.9300000 -1.5820000
## t-value — -10.52099 -15.386196
## df — 86.53800 76.51587
## p-value — < .0000001 < .0000001
##
## versicolor Mean difference — -0.6520000
## t-value — -5.629165
## df — 94.02549
## p-value — 0.0000006
##
## virginica Mean difference —
## t-value —
## df —
## p-value —
## --------------------------------------------------------------------------
## Note. * p < .05, ** p < .01, *** p < .001
利用R语言对数据进行可视化
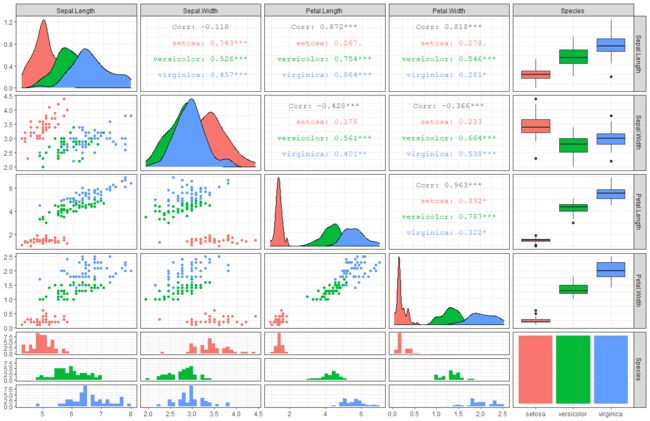
到了这里,事情解决了,对么?很传统的方法,得到了想知道的一切。数据有差异性,完美。但其实,R语言能够做的更多。我更愿意称R语言为可视化统计检验方法。相比于传统方法的先检验差异性再作图,R语言是先做可视化再做具体的方差分析。例如,我们可以一行代码查看数据的关系。这里面包含了数据的相关散点图,直方图,相关系数和箱线图。依靠这些你可以更好的提出你需要的科学假设。
ggpairs(data, mapping = aes(color = Species)) + theme_bw()
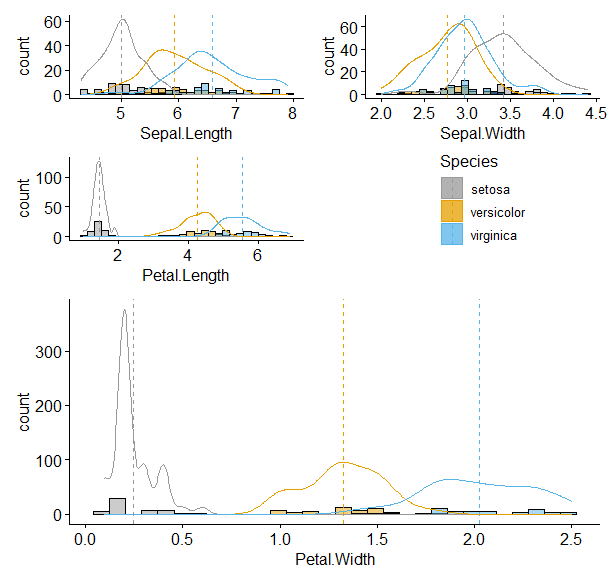
通过直方图可以很容易看到数据的分布情况。是否满足正态还有方差齐性是不是变得更加具象了?
p_SL <- data %>% gghistogram(data = ., x= 'Sepal.Length', add = 'mean', fill = 'Species', palette = cbPalette,add_density = TRUE, legend = 'right')
p_SW <- data %>% gghistogram(data = ., x= 'Sepal.Width', add = 'mean', fill = 'Species', palette = cbPalette,add_density = TRUE, legend = 'right')
p_PL <- data %>% gghistogram(data = ., x= 'Petal.Length', add = 'mean', fill = 'Species', palette = cbPalette,add_density = TRUE, legend = 'right')
p_PW <- data %>% gghistogram(data = ., x= 'Petal.Width', add = 'mean', fill = 'Species', palette = cbPalette,add_density = TRUE, legend = 'none' )
(p_SL + p_SW + p_PL + guide_area() + plot_layout(guides = 'collect')) /p_PW
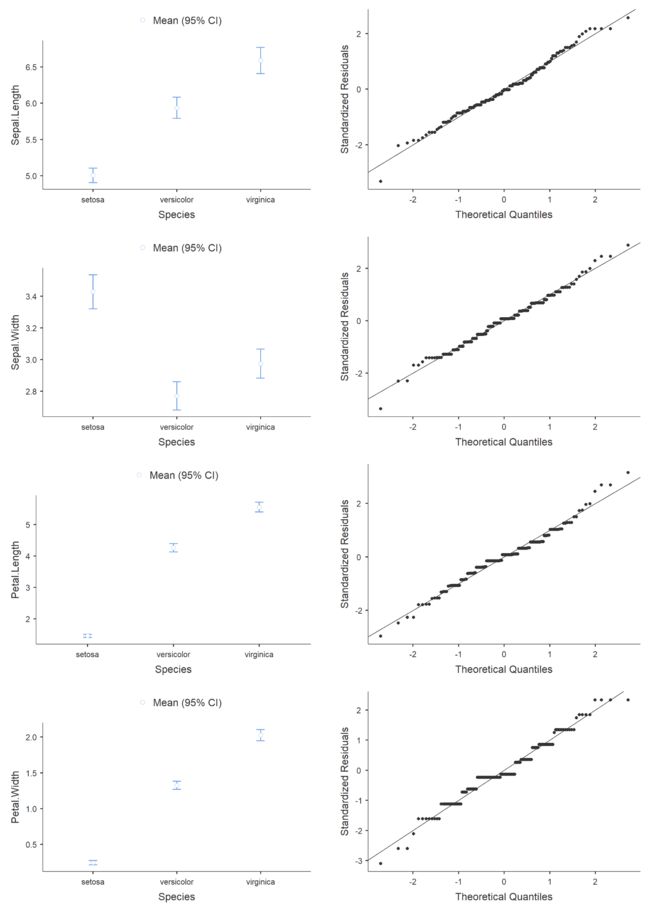
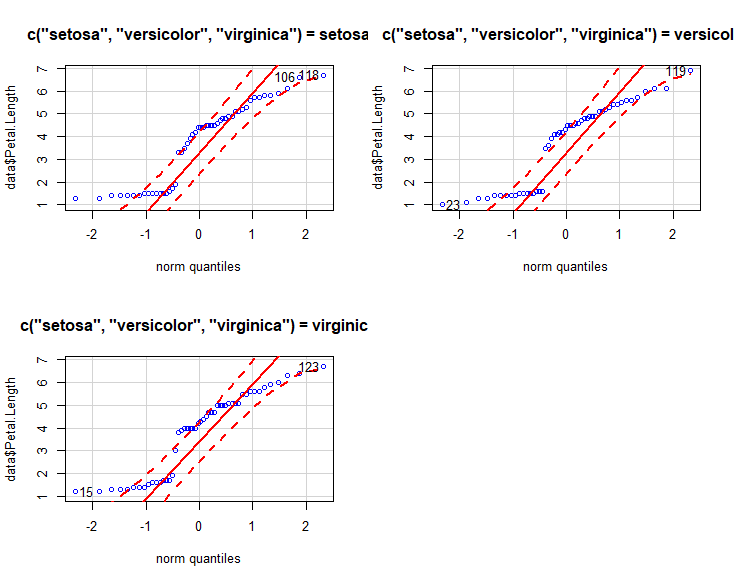
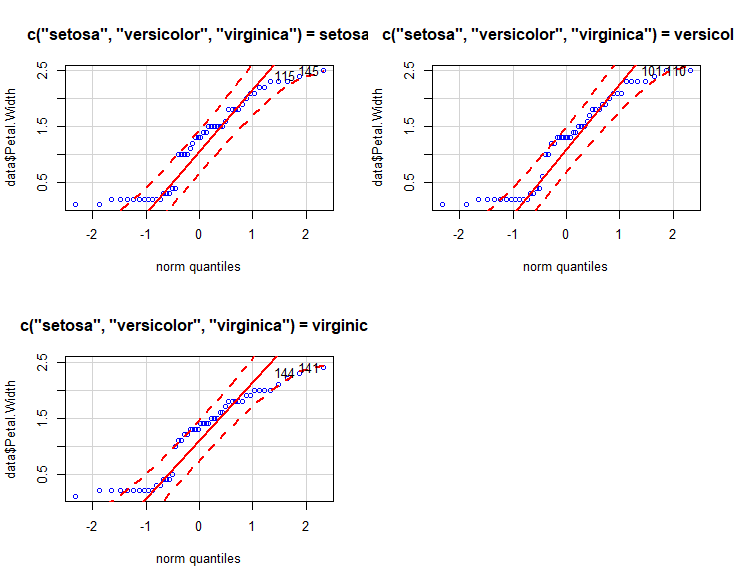
跑个qq图,配合置信区间。
qqPlot(data$Sepal.Length ,main="qq plot", groups = c("setosa","versicolor","virginica"), col="blue", col.lines="red")
qqPlot(data$Sepal.Width ,main="qq plot", groups = c("setosa","versicolor","virginica"), col="blue", col.lines="red")
qqPlot(data$Petal.Length ,main="qq plot", groups = c("setosa","versicolor","virginica"), col="blue", col.lines="red")
qqPlot(data$Petal.Width ,main="qq plot", groups = c("setosa","versicolor","virginica"), col="blue", col.lines="red")
然后是数据结果的分组可视化。
compare_group <- list(c('setosa','versicolor'),c('versicolor','virginica'),c('setosa','virginica'))
p1 <- ggboxplot(data, x = 'Species', y = 'Sepal.Length', add = c('jitter','mean_se'), color = 'Species', palette = cbPalette) +
stat_compare_means(comparisons = compare_group, label = 'p.signif',method = 'wilcox.test')
p2 <- ggboxplot(data, x = 'Species', y = 'Sepal.Width', add = c('jitter','mean_se'), color = 'Species', palette = cbPalette) +
stat_compare_means(comparisons = compare_group, label = 'p.signif',method = 'wilcox.test')
p3 <- ggboxplot(data, x = 'Species', y = 'Petal.Length', add = c('jitter','mean_se'), color = 'Species', palette = cbPalette) +
stat_compare_means(comparisons = compare_group, label = 'p.signif',method = 'wilcox.test')
p4 <- ggboxplot(data, x = 'Species', y = 'Petal.Width', add = c('jitter','mean_se'), color = 'Species', palette = cbPalette) +
stat_compare_means(comparisons = compare_group, label = 'p.signif',method = 'wilcox.test')
p1 + p2 + p3 + p4 + plot_layout(guides = 'collect') & theme(legend.position = 'bottom')
还可以换个炫酷的效果。这里用的Games_Howell test。不喜欢可以换回t检验,大家可以自己试试看。
p1 <- data %>% group_by(Species) %>% ggstatsplot::ggbetweenstats(x = Species, y = Sepal.Length, nboot = 10, messages = FALSE)
p2 <- data %>% group_by(Species) %>% ggstatsplot::ggbetweenstats(x = Species, y = Sepal.Width, nboot = 10, messages = FALSE)
p3 <- data %>% group_by(Species) %>% ggstatsplot::ggbetweenstats(x = Species, y = Petal.Length, nboot = 10, messages = FALSE)
p4 <- data %>% group_by(Species) %>% ggstatsplot::ggbetweenstats(x = Species, y = Petal.Width, nboot = 10, messages = FALSE)
p1 + p2 + p3 + p4
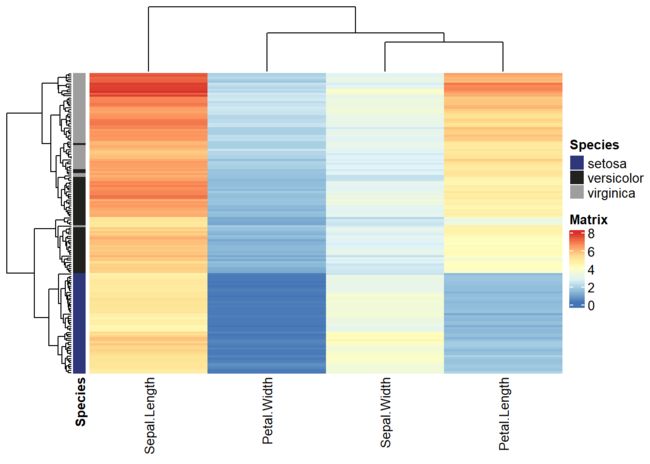
最后我们可以用heatmap查看各个数据在四个维度上的表现,这里我因为同时装了两个包所以,冲突了。所以需要申明到底用的是哪个包的pheatmap。
set.seed(2020)
anno <- data.frame(Species = data$Species)
row.names(anno) <- row.names(data)
ComplexHeatmap::pheatmap(data[,-5], border_color = gpar(col = "black", lty = 2), cluster_rows = T, cluster_cols = T, show_rownames = F, show_colnames = T, annotation_row = anno)
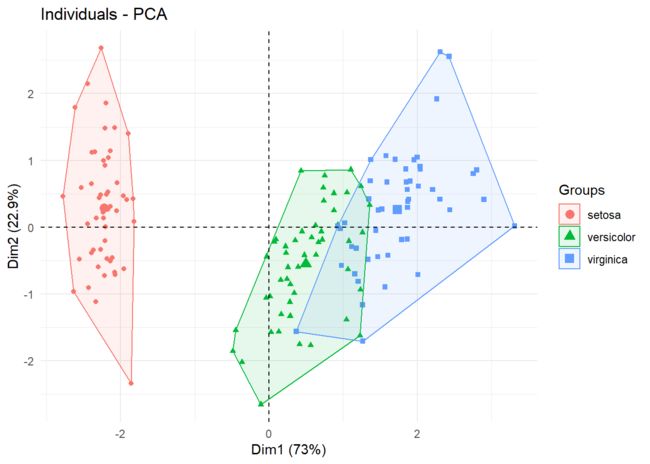
最后根据四个petal和sepal的长宽数据,我们做个pca分类图。
data.pca <- PCA(data[,-5], graph = FALSE)
fviz_pca_ind(data.pca,
geom.ind = "point",
col.ind = data$Species,
addEllipses = TRUE,
ellipse.type = 'convex',
legend.title = "Groups"
) + theme_minimal()
row.names(data) <- paste0("row_", seq(nrow(data)))
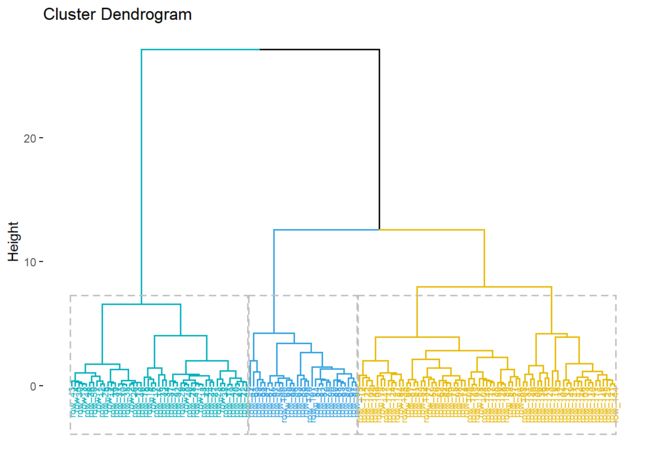
res <- factoextra::hcut(data[,-5], k = 3, stand = TRUE)
fviz_dend(res, rect = TRUE, cex = 0.5,
k_colors = c("#00AFBB","#2E9FDF", "#E7B800", "#FC4E07"))
Tidymodel建模
到这里,我们体验了大部分统计可视化的内容。那么R还能做什么?既然我们知道所谓统计分析本身就是建模,那么我们能否用更复杂的模型对petal和sepal的长宽等因子进行重要性检验呢?所以又有了这部分,利用tidymodels的建模,并根据模型对数据分类。
library('tidymodels')
library('caret')
#固定随机数,方便复现
set.seed(2020)
下面是整个建模的流程
#将数据分成3份两份作为训练数据,一份作为测试准确性数据。
data_split <- initial_split(data,prop=.66)
data_train <- training(data_split)
data_test <-testing(data_split)
#bootstrap创造一个数据用来tuning模型
Spec_boost <- bootstraps(data_train, times = 30)
#设定模型数据转换,在这里可以做中心化,标准化,数据合并等一系列操作。最后传入的prep记录所有的操作。另外在这里Species~.意思是Species作为分类结果,其他几列作为predictor预测分类。
data_rec <-
recipe(Species ~.,data = data_train) %>%
prep()
#查看训练的数据,这里用juice很形象的说你的数据是榨出来的。但这里没对表型数据做过多调整。
juice(data_rec)
#设定模型,这里mtry和min_n使用tune函数调整。模式选择分类,随机森林的引擎选择randomForest,另外还有一个引擎叫ranger。具体到后面会有细微差别。
rf_model<-rand_forest(mtry=tune(),min_n = tune())%>%
set_mode("classification")%>%
set_engine("randomForest")
#设定工作流,主要是使用哪个模型以及使用哪个数据方便后期做具体的调参。
rf_wflow <-
workflow() %>%
add_recipe(data_rec)%>%
add_model(rf_model)
#激活平行线程
doParallel::registerDoParallel()
#使用tune_grid函数和boostrap的数据进行大致的调参。
rf_results <-
rf_wflow %>%
tune_grid(resamples = Spec_boost)
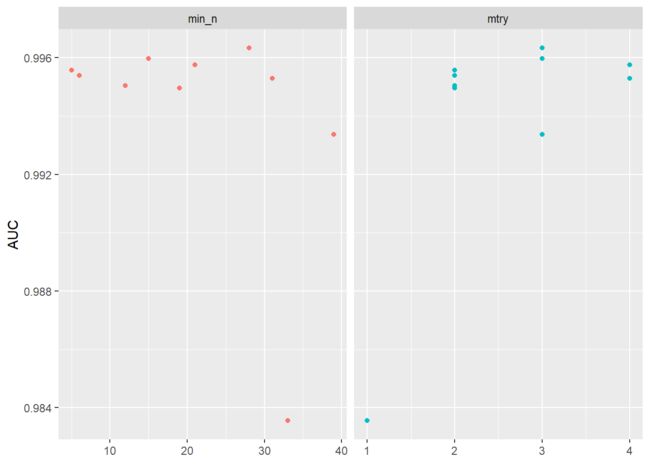
#参数打分可视化
rf_results %>%
collect_metrics() %>%
filter(.metric == "roc_auc") %>%
select(mean, min_n, mtry) %>%
pivot_longer(min_n:mtry,
values_to = "value",
names_to = "parameter"
) %>%
ggplot(aes(value, mean, color = parameter)) +
geom_point(show.legend = FALSE) +
facet_wrap(~parameter, scales = "free_x") +
labs(x = NULL, y = "AUC")
#选择最佳模型参数
so_best <-
rf_results %>%
select_best(metric = "roc_auc")
#构建模型
rf_fit <- rand_forest(mode = 'classification',
mtry = so_best$mtry,
min_n = so_best$min_n) %>%
set_engine("randomForest") %>%
fit( Species ~ ., data = data_train)
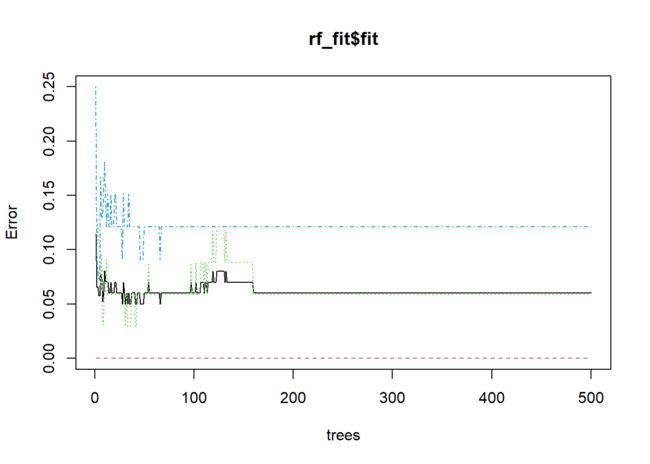
plot(rf_fit$fit)
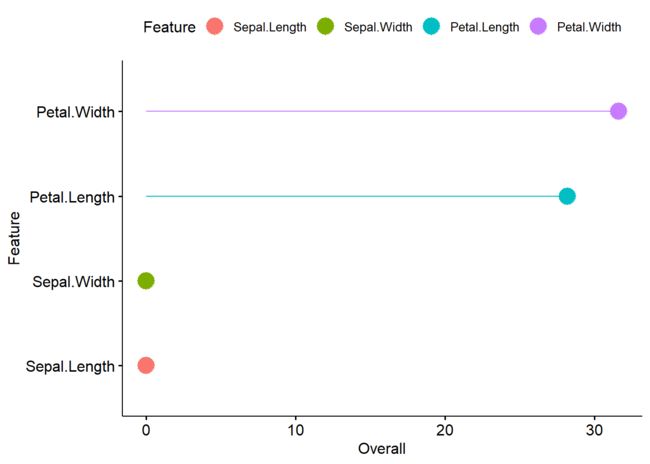
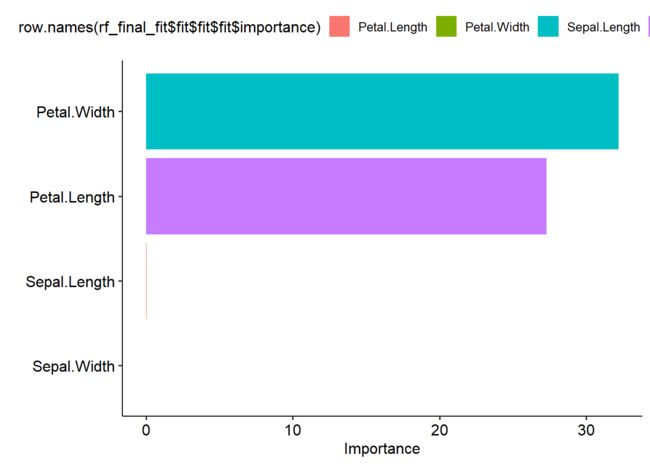
#各因子重要性可视化
Importance <- varImp(rf_fit$fit)
Importance$Feature <- row.names(Importance)
Importance
## Overall Feature
## Sepal.Length 0.007138445 Sepal.Length
## Sepal.Width 0.000000000 Sepal.Width
## Petal.Length 28.186545520 Petal.Length
## Petal.Width 31.611573933 Petal.Width
ggdotchart(Importance,x = 'Feature', y = 'Overall', color = 'Feature',
palette = 'mpg', sorting = "descending",
font.label = list(color = "white", size = 9, vjust = 0.5),add.params = list(color = "Feature"), add = "segments", rotate = TRUE, group = "Feature", dot.size = 6,
ggtheme = theme_pubr())
#激活平行线程
doParallel::registerDoParallel()
#设定参数
rf_grid <- grid_regular(
mtry(range = c(1,4)),
min_n(range = c(10,30)),
levels = 10
)
#调参结果
regular_res <- rf_wflow %>%
tune_grid(
resamples = Spec_boost,
grid = rf_grid
)
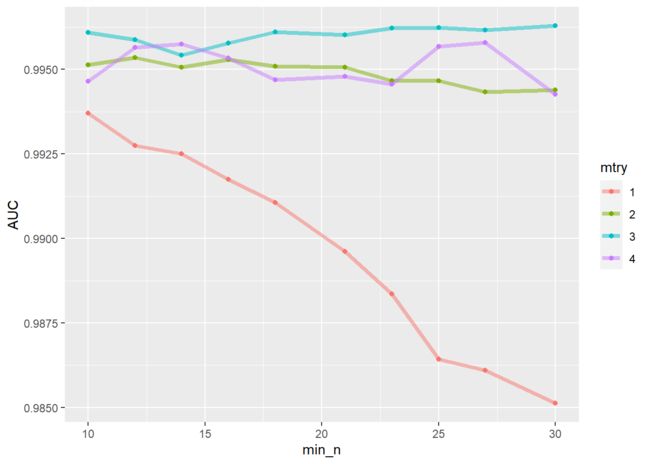
#结果可视化
regular_res%>%
collect_metrics() %>%
filter(.metric == "roc_auc") %>%
mutate(mtry = factor(mtry)) %>%
ggplot(aes(min_n, mean, color = mtry)) +
geom_line(alpha = 0.5, size = 1.5) +
geom_point() +
labs(y = "AUC")
#调参后通过roc_auc指标选择最好的模型参数
so_best <-
regular_res %>%
select_best(metric = "roc_auc")
#使用最佳参数去构建模型
rf_final_fit<-rf_wflow%>%
finalize_workflow(so_best)%>%
fit(data = data_train)
#设定好数据调整的方法(还记得recipes么)
data_rec3 <-rf_final_fit%>%
pull_workflow_prepped_recipe()
#用同样方法去变换test数据集,然后用模型拟合test数据集,输出结果。
rf_final_fit%>%
pull_workflow_fit()%>%
predict( new_data = bake(data_rec3, data_test))%>%
bind_cols(data_test, .)%>%
metrics(truth = Species, estimate =.pred_class)
## # A tibble: 2 x 3
## .metric .estimator .estimate
##
## 1 accuracy multiclass 0.96
## 2 kap multiclass 0.940
#查看参数重要性
vi(rf_final_fit$fit$fit)
vip(rf_final_fit$fit$fit, mapping = aes(fill = row.names(rf_final_fit$fit$fit$fit$importance))) + theme_pubr()