BraTS数据集处理详解(附代码详解)
代码参考:https://github.com/sinclairjang/3D-MRI-brain-tumor-segmentation-using-autoencoder-regularization
数据集来源:BraTs 2018
参考论文:https://arxiv.org/abs/1810.11654
3D-MRI-brain-tumor-segmentation-using-autoencoder-regularization
参考作者数据集预处理方式,后续模型将持续更新…
目录
-
- 数据集详解
- 数据集介绍
- 数据集处理(附代码详解)
- 一. data load
- 二. 图像预处理
- 1. 数据预处理
- 二.生成data,label
- 总结
数据集详解
- BraTS 数据集是脑肿瘤分割比赛数据集,brats 2018中的训练集( training set) 有285个病例,每个病例有四个模态(t1、t2、flair、t1ce),需要分割三个部分:whole tumor(WT), enhance tumor(ET), and tumor core(TC).
- t1、t2、flair、t1ce可以理解为核磁共振图像的四个不同纬度信息,每个序列的图像shape为(155,240,240)
- 目标是分割出三个label。对应医学中的三个不同肿瘤类型。
- 以上是本人理解,非医学专业,如有错误欢迎指正。
数据集介绍
- BraTs数据集类型为XX.nii.gz,分别对应t1、t2、flair、t1ce,seg,其中seg是分割图像。图像大小均为(155,240,240)
数据集处理(附代码详解)
一. data load
t1 = glob.glob(r'/home/locco/brats17/MICCAI_BraTS_2019_Data_Training/MICCAI_BraTS_2019_Data_Training/*/*/*t1.nii.gz')
t2 = glob.glob(r'/home/locco/brats17/MICCAI_BraTS_2019_Data_Training/MICCAI_BraTS_2019_Data_Training/*/*/*t2.nii.gz')
flair = glob.glob(r'/home/locco/brats17/MICCAI_BraTS_2019_Data_Training/MICCAI_BraTS_2019_Data_Training/*/*/*flair.nii.gz')
t1ce = glob.glob(r'/home/locco/brats17/MICCAI_BraTS_2019_Data_Training/MICCAI_BraTS_2019_Data_Training/*/*/*t1ce.nii.gz')
seg = glob.glob(r'/home/locco/brats17/MICCAI_BraTS_2019_Data_Training/MICCAI_BraTS_2019_Data_Training/*/*/*seg.nii.gz')
分别生成t1、t2、flair、t1ce、seg对应文件目录
利用SimpleITK读取图像
def read_img(img_path):
return sitk.GetArrayFromImage(sitk.ReadImage(img_path))
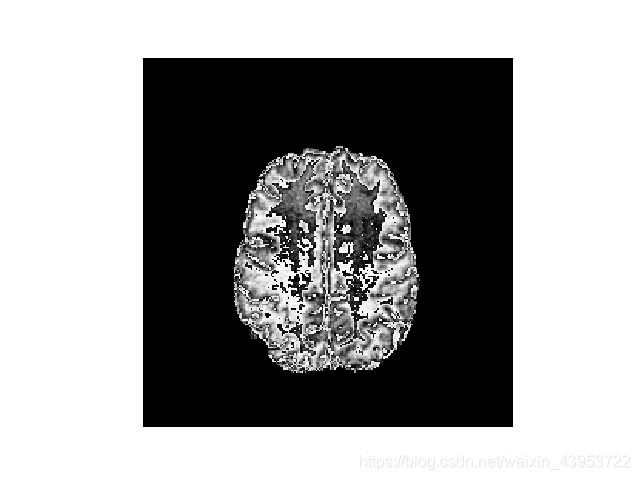
读取图像,并可视化(可视化的是三维图像中的一个切片图像)
import matplotlib.pyplot as plt
img = (read_img(t1[0])[100]).astype(np.uint8)
plt.imshow(img)
二. 图像预处理
1. 数据预处理
- 将原图像压缩为(80,96,64)大小图像,并将4个序列图像放在统一纬度,所以经过数据处理后的每个样本图像的维度为(4,80,96,64)
- 同样将mask图像作相同处理,由于分割图像的label包含3个纬度信息(3个不同的肿瘤类型),所以处理后的mask纬度为(3,80,96,64)
def resize(img, shape, mode='constant', orig_shape=(155, 240, 240)):
"""
Wrapper for scipy.ndimage.zoom suited for MRI images.
"""
assert len(shape) == 3, "Can not have more than 3 dimensions"
factors = (
shape[0]/orig_shape[0],
shape[1]/orig_shape[1],
shape[2]/orig_shape[2]
)
# Resize to the given shape
return zoom(img, factors, mode=mode)
def preprocess(img, out_shape=None):
"""
Preprocess the image.
Just an example, you can add more preprocessing steps if you wish to.
"""
if out_shape is not None:
img = resize(img, out_shape, mode='constant')
# Normalize the image
mean = img.mean()
std = img.std()
return (img - mean) / std
def preprocess_label(img, out_shape=None, mode='nearest'):
"""
Separates out the 3 labels from the segmentation provided, namely:
GD-enhancing tumor (ET — label 4), the peritumoral edema (ED — label 2))
and the necrotic and non-enhancing tumor core (NCR/NET — label 1)
"""
ncr = img == 1 # Necrotic and Non-Enhancing Tumor (NCR/NET)
ed = img == 2 # Peritumoral Edema (ED)
et = img == 4 # GD-enhancing Tumor (ET)
if out_shape is not None:
ncr = resize(ncr, out_shape, mode=mode)
ed = resize(ed, out_shape, mode=mode)
et = resize(et, out_shape, mode=mode)
return np.array([ncr, ed, et], dtype=np.uint8)
二.生成data,label
input_shape = (4, 80, 96, 64)
output_channels = 3
data = np.empty((len(data_paths),) + input_shape, dtype=np.float32)
labels = np.empty((len(data_paths), output_channels) + input_shape[1:], dtype=np.uint8)
import math
# Parameters for the progress bar
total = len(data_paths)
step = 25 / total
for i, imgs in enumerate(data_paths):
try:
data[i] = np.array([preprocess(read_img(imgs[m]), input_shape[1:]) for m in ['t1', 't2', 't1ce', 'flair']], dtype=np.float32)
labels[i] = preprocess_label(read_img(imgs['seg']), input_shape[1:])[None, ...]
# Print the progress bar
print('\r' + f'Progress: '
f"[{'=' * int((i+1) * step) + ' ' * (24 - int((i+1) * step))}]"
f"({math.ceil((i+1) * 100 / (total))} %)",
end='')
except Exception as e:
print(f'Something went wrong with {imgs["t1"]}, skipping...\n Exception:\n{str(e)}')
continue
总结
本文仅po出BraTs数据集加载与预处理部分,刚好本人最近再看医学图像分割方面的论文。由于数据集为3D MRI数据集,处理时还需要大家了解一定的医学图象知识。
作者原文中采用了一种改良的U-Net网络进行训练,后续将更新模型部分详解,欢迎交流。