吴恩达机器学习K-Mean算法和PCA算法的MATLAB实现(对应ex7练习)
K-mean算法
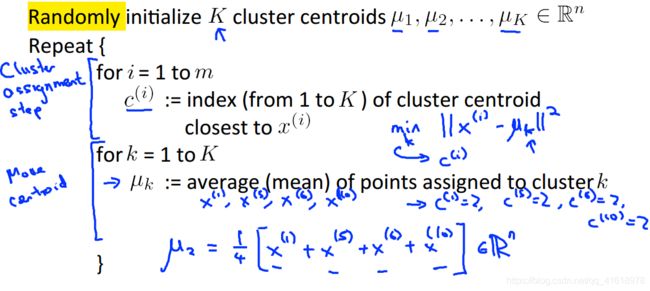
前言:K-mean算法的思想和实现都不难,整个算法主要分为两步:1、找到与每个样本距离最近的质心,将样本与最近的质心关联起来;2、根据每个质心的关联样本,重新计算质心的位置。下面是吴恩达课件里的说明,说的十分清晰。
在实现K-mean算法的过程中,质心的初始位置一般是随机选取样本点作为质心。例如需要将样本分为K类,那么从样本中随机选取K个样本作为初始化质心即可。样本的初始位置选取与最终的分类效果有很大的关系。
下面给出作业中的关键代码。
findClosestCentroids.m
这个函数实现的功能是找到距离每个样本点最近的质心,并将质心的序号用一个矩阵存储起来,便于之后重新计算质心的位置使用。如下图给出的解释,其中c(i)和idx(i)是一样的,都代表距离第i个样本点最近的质心的序号。
因此,这个函数的主要实现是一个距离计算并求最小值的过程。下面给出MATLAB实现代码。
function idx = findClosestCentroids(X, centroids)
%FINDCLOSESTCENTROIDS computes the centroid memberships for every example
% idx = FINDCLOSESTCENTROIDS (X, centroids) returns the closest centroids
% in idx for a dataset X where each row is a single example. idx = m x 1
% vector of centroid assignments (i.e. each entry in range [1..K])
%
% Set K
K = size(centroids, 1);%质心的数量
% You need to return the following variables correctly.
idx = zeros(size(X,1), 1);%用于存储每个点最近的质心的序号
% ====================== YOUR CODE HERE ======================
% Instructions: Go over every example, find its closest centroid, and store
% the index inside idx at the appropriate location.
% Concretely, idx(i) should contain the index of the centroid
% closest to example i. Hence, it should be a value in the
% range 1..K
%
% Note: You can use a for-loop over the examples to compute this.
%
val = zeros(K,1);%创建一个变量用于存储每个中心点到数据点的距离,之后取其中最小的
for i = 1 : size(X,1);
for j = 1 : size(centroids,1);
val(j) = sum((X(i,:) - centroids(j,:)) .^ 2);%样本点与每个质心的距离
end
[~,idx(i)] = min(val);%取出最近的质心的序号,这里~的作用是占位符
end
% =============================================================
endcomputeCentroids.m
这个函数实际就是K-mean算法的第二步,根据第一步得到的集群重新计算每个质心的位置。具体实现是找到与每个质心最近的点,将这些点求和之后取平均,这个均值就是新的质心位置。在MATLAB中的实现过程如下。
function centroids = computeCentroids(X, idx, K)
%COMPUTECENTROIDS returns the new centroids by computing the means of the
%data points assigned to each centroid.
% centroids = COMPUTECENTROIDS(X, idx, K) returns the new centroids by
% computing the means of the data points assigned to each centroid. It is
% given a dataset X where each row is a single data point, a vector
% idx of centroid assignments (i.e. each entry in range [1..K]) for each
% example, and K, the number of centroids. You should return a matrix
% centroids, where each row of centroids is the mean of the data points
% assigned to it.
%
% Useful variables
[m n] = size(X);%
% You need to return the following variables correctly.
centroids = zeros(K, n);%
% ====================== YOUR CODE HERE ======================
% Instructions: Go over every centroid and compute mean of all points that
% belong to it. Concretely, the row vector centroids(i, :)
% should contain the mean of the data points assigned to
% centroid i.
%
% Note: You can use a for-loop over the centroids to compute this.
%
for i = 1 : K;
num = find(idx == i);%找到与第i个中心点相近的所有点,num里存储的是每个点在X中的行序号
centroids(i,:) = sum(X(num,:)) ./ size(num,1);%对这些点求和之后取平均值并更新中心点的坐标
end
% =============================================================
end
在完成上面两步之后,剩下的就是迭代过程,通过一定次数的迭代求解质心的位置,最后将迭代完成的质心用来找出离每个质心最近的样本点(也就是第一步的过程),并将其分为一个集群既可。
PCA算法
前言:PCA算法的难点主要是在推导和数学理解上,对于其实现并不难。对于PCA算法的MATLAB实现,同样总结为两步:1、数据的预处理,根据吴恩达视频中的讲解,需要将样本的特征去中心化,使样本特征的和值为。具体实现是求出每个特征的均值,用原来的特征减去均值代替即可(对于不同的特征可能还需要除样本的标准差来进行缩放);
2、计算样本的协方差并进行特征值和特征向量的分解(可以借助MATLAB中的svd函数实现),将特征向量中的前k列取出与原样本做计算即可。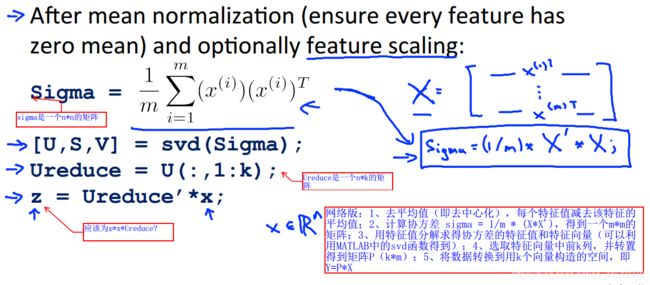
接下来是作业中有关的代码片段。
featureNormalize.m
function [X_norm, mu, sigma] = featureNormalize(X)
%FEATURENORMALIZE Normalizes the features in X
% FEATURENORMALIZE(X) returns a normalized version of X where
% the mean value of each feature is 0 and the standard deviation
% is 1. This is often a good preprocessing step to do when
% working with learning algorithms.
mu = mean(X);
X_norm = bsxfun(@minus, X, mu);%对两个矩阵A和B之间的每一个元素进行指定的计算(函数fun指定)
%该函数的具体实现过程是:
%①判断A和B的维度是否相同,如果相同,直接进行操作;
%②如果A和B的维度不同,则A或者B必须有一个在某个维度上是1,比如,上例中的mu在行方向维度是1,那么,
%该函数将会对行向量mu在行方向上进行复制,使其与矩阵X具有相同的行维度,然后,再进行X-mu维度扩充后的矩阵
sigma = std(X_norm);%计算标准差
X_norm = bsxfun(@rdivide, X_norm, sigma);
% ============================================================
end
下面两个m文件其实做的就是第二步,合在一起看便于理解。
pca.m
function [U, S] = pca(X)
%PCA Run principal component analysis on the dataset X
% [U, S, X] = pca(X) computes eigenvectors of the covariance matrix of X
% Returns the eigenvectors U, the eigenvalues (on diagonal) in S
%
% Useful values
[m, n] = size(X);
% You need to return the following variables correctly.
U = zeros(n);
S = zeros(n);
% ====================== YOUR CODE HERE ======================
% Instructions: You should first compute the covariance matrix. Then, you
% should use the "svd" function to compute the eigenvectors
% and eigenvalues of the covariance matrix.
%
% Note: When computing the covariance matrix, remember to divide by m (the
% number of examples).
%
sigma = 1 / m * X' * X;%n*n
[U,S,~] = svd(sigma);
% =========================================================================
end
projectData.m
function Z = projectData(X, U, K)
%PROJECTDATA Computes the reduced data representation when projecting only
%on to the top k eigenvectors
% Z = projectData(X, U, K) computes the projection of
% the normalized inputs X into the reduced dimensional space spanned by
% the first K columns of U. It returns the projected examples in Z.
%
% You need to return the following variables correctly.
Z = zeros(size(X, 1), K);
% ====================== YOUR CODE HERE ======================
% Instructions: Compute the projection of the data using only the top K
% eigenvectors in U (first K columns).
% For the i-th example X(i,:), the projection on to the k-th
% eigenvector is given as follows:
% x = X(i, :)';
% projection_k = x' * U(:, k);
%
Ureduce = U(:, 1:K);
Z = X*Ureduce;
%课件版
% sigma = 1 / size(X,1) * (X' * X);
% [U,S,~] = svd(sigma);%这里得到的U是协方差sigma的特征向量按列排列组成,S为对角线矩阵,其值是由大到小排列的特征值
% Ureduce = U(:,1:K);%n * k
% Z = X * Ureduce;
% =============================================================
end最后一步的数据的还原,只要知道降维怎么做,之后再恢复数据就很容易。
recoverData.m
function X_rec = recoverData(Z, U, K)
%RECOVERDATA Recovers an approximation of the original data when using the
%projected data
% X_rec = RECOVERDATA(Z, U, K) recovers an approximation the
% original data that has been reduced to K dimensions. It returns the
% approximate reconstruction in X_rec.
%
% You need to return the following variables correctly.
X_rec = zeros(size(Z, 1), size(U, 1));
% ====================== YOUR CODE HERE ======================
% Instructions: Compute the approximation of the data by projecting back
% onto the original space using the top K eigenvectors in U.
%
% For the i-th example Z(i,:), the (approximate)
% recovered data for dimension j is given as follows:
% v = Z(i, :)';
% recovered_j = v' * U(j, 1:K)';
%
% Notice that U(j, 1:K) is a row vector.
%
Ureduce = U(:, 1:K);
X_rec=Z*Ureduce';
% =============================================================
end