使用机器学习算法实现单细胞测序数据的降维及聚类(一)
主要代码参考于此,感谢b站大学
主要代码参考于此,感谢GitHub老师
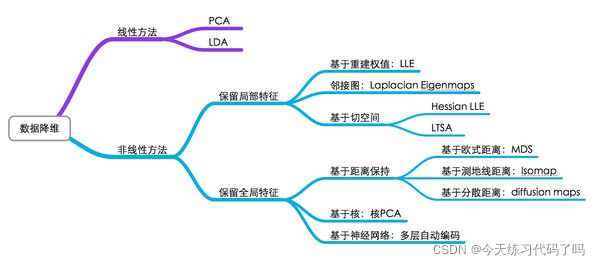
本篇主要记录一下几种常用的降维算法
数据集和文中代码可从我的gitee中中获取
数据是darmanis数据集,包括466个细胞2000个高表达量基因,分为九种类型的细胞集群。数据部分截图:
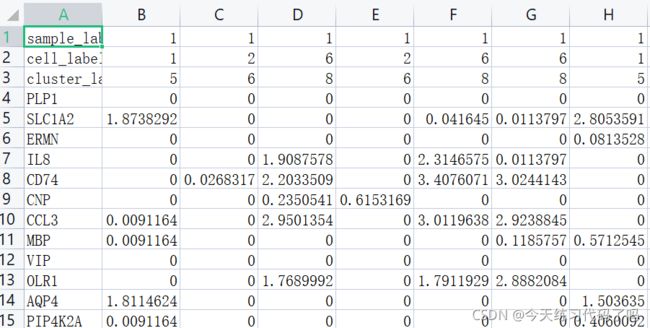
其中行为基因列为细胞,每个数据表示基因在细胞中的表达量。
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
from sklearn.preprocessing import StandardScaler, MinMaxScaler
def pca(data, n_dim):
num_cells, num_genes = data.shape
# 去均值
data = data - np.mean(data, axis=0)
# 获得协方差矩阵 Conv_data=(num_genes,num_genes)
Conv_data = np.dot(data.T, data) / num_cells - 1
# 求特征值和特征向量
eig_values, eig_vectors = np.linalg.eig(Conv_data)
# 将特征值从大到小排序,并取前n_dim个
indexs = np.argsort(-eig_values)[:n_dim]
# 取对应的特征向量组成降维矩阵 pick_eig_vector=(num_genes,n_dim)
pick_eig_vector = eig_vectors[:, indexs]
# 获得新的矩阵 New_data=(num_cells,n_dim)
New_data = np.dot(data, pick_eig_vector)
return New_data
def draw_pic(datas, labs):
plt.cla()
unque_labs = np.unique(labs)
colors = [plt.cm.Spectral(each)
for each in np.linspace(0, 1, len(unque_labs))]
p = []
legends = []
for i in range(len(unque_labs)):
index = np.where(labs == unque_labs[i])
pi = plt.scatter(datas[index, 0], datas[index, 1], c=[colors[i]])
p.append(pi)
legends.append(unque_labs[i])
plt.legend(p, legends)
plt.show()
if __name__ == "__main__":
data = pd.read_csv('dataset/darmanis.csv')
data = data.to_numpy().T
matrix = data[1:, 2:].astype(float)
print(matrix.shape)
labels = data[1:,1:2].astype(int).flatten()
print(labels.shape)
new_data = pca(matrix, 2)
draw_pic(new_data, labels)
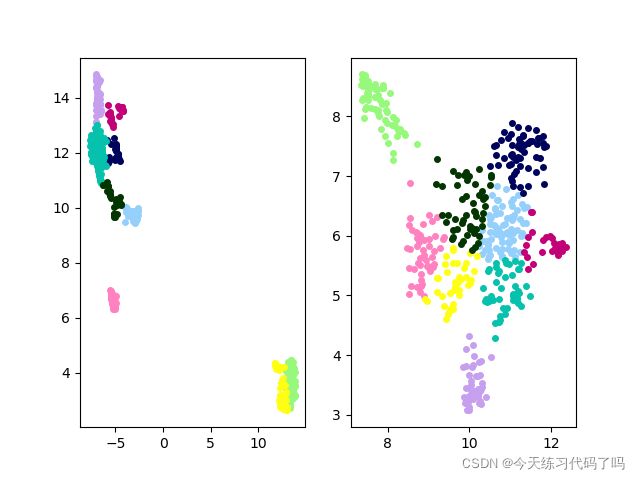
降维效果如图
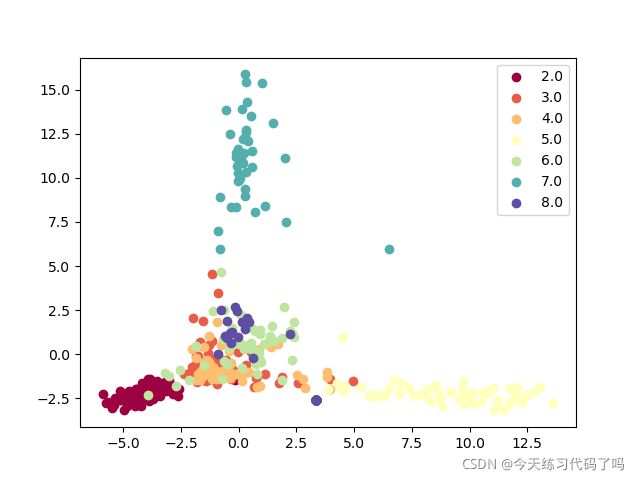
优点
仅仅需要以方差衡量信息量,不受数据集外的因素影响.
各主成分之间正交,可消除原始数据成分间的相互影响的因素
计算方法简单,主要运算是特征值分解,易于实现
缺点
主成分各个特征维度的含义具有一定的模糊性,不如原始样本特征的解释性强
方差小的非主成分也可能含有对样本差异的重要信息,因维度丢弃可能对后续数据处理有影响
详细原理参考:https://blog.csdn.net/andyjkt/article/details/107914007
2.T-SNE
T-SNE是目前来说效果最好的数据降维与可视化方法,但是它的缺点也很明显,比如:占内存大,运行时间长。但是,当我们想要对高维数据进行分类,又不清楚这个数据集有没有很好的可分性(即同类之间间隔小,异类之间间隔大),可以通过t-SNE投影到2维或者3维的空间中观察一下。如果在低维空间中具有可分性,则数据是可分的;如果在高维空间中不具有可分性,可能是数据不可分,也可能仅仅是因为不能投影到低维空间。
t-SNE(TSNE)将数据点之间的相似度转换为概率。原始空间中的相似度由高斯联合概率表示,嵌入空间的相似度由“student-t分布”表示。
t-SNE主要是关注数据的局部结构。
TSNE通过原始空间和嵌入空间的联合概率的Kullback-Leibler(KL)散度来评估可视化效果的好坏,也就是说用有关KL散度的函数作为loss函数,然后通过梯度下降最小化loss函数,最终获得收敛结果。注意,该loss不是凸函数,即具有不同初始值的多次运行将收敛于KL散度函数的局部最小值中,以致获得不同的结果。因此,尝试不同的随机数种子(Python中可以通过设置seed来获得不同的随机分布)有时候是有用的,并选择具有最低KL散度值的结果。
使用t-SNE的缺点大概是:
1.t-SNE的计算复杂度很高,在数百万个样本数据集中可能需要几个小时,而PCA可以在几秒钟或几分钟内完成.
2.算法是随机的,具有不同种子的多次实验可以产生不同的结果。虽然选择loss最小的结果就行,但可能需要多次实验以选择超参数。
3.全局结构未明确保留。这个问题可以通过PCA初始化点(使用init =‘pca’)来缓解。
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
from PCA import pca
from K_Means import KMeans
# 计算任意两点之前距离 ||x_i-x_j||^2
# X 维度 [N,D]
def cal_pairwise_dist(X):
sum_X = np.sum(np.square(X), 1)
D = np.add(np.add(-2 * np.dot(X, X.T), sum_X).T, sum_X)
# 返回任意两个点之间距离
return D
# 计算P_ij 以及 log松弛度
def calc_P_and_entropy(D, beta=1.0):
P = np.exp(-D.copy() * beta)
sumP = np.sum(P)
# 计算熵
log_entropy = np.log(sumP) + beta * np.sum(D * P) / sumP
P = P / sumP
return P, log_entropy
# 二值搜索寻找最优的 sigma
def binary_search(D, init_beta, logU, tol=1e-5, max_iter=50):
beta_max = np.inf
beta_min = -np.inf
beta = init_beta
P, log_entropy = calc_P_and_entropy(D, beta)
diff_log_entropy = log_entropy - logU
m_iter = 0
while np.abs(diff_log_entropy) > tol and m_iter < max_iter:
# 交叉熵比期望值大,增大beta
if diff_log_entropy > 0:
beta_min = beta
if beta_max == np.inf or beta_max == -np.inf:
beta = beta * 2
else:
beta = (beta + beta_max) / 2.
# 交叉熵比期望值小, 减少beta
else:
beta_max = beta
if beta_min == -np.inf or beta_min == -np.inf:
beta = beta / 2
else:
beta = (beta + beta_min) / 2.
# 重新计算
P, log_entropy = calc_P_and_entropy(D, beta)
diff_log_entropy = log_entropy - logU
m_iter = m_iter + 1
# 返回最优的 beta 以及所对应的 P
return P, beta
# 给定一组数据 datas :[N,D]
# 计算联合概率 P_ij : [N,N]
def p_joint(datas, target_perplexity):
N, D = np.shape(datas)
# 计算两两之间的距离
distances = cal_pairwise_dist(datas)
beta = np.ones([N, 1]) # beta = 1/(2*sigma^2)
logU = np.log(target_perplexity)
p_conditional = np.zeros([N, N])
# 对每个样本点搜索最优的sigma(beta) 并计算对应的P
for i in range(N):
if i % 500 == 0:
print("给 %d 个点计算联合概率 P_ij" % (i))
# 删除 i -i 点
Di = np.delete(distances[i, :], i)
# 进行二值搜索,寻找 beta
# 使 log_entropy 最接近 logU
P, beta[i] = binary_search(Di, beta[i], logU)
# 在ii的位置插0
p_conditional[i] = np.insert(P, i, 0)
# 计算联合概率
P_join = p_conditional + p_conditional.T
P_join = P_join / np.sum(P_join)
print("σ的平均值: %f" % np.mean(np.sqrt(1 / beta)))
return P_join
# Y : 低维数据 [N,d]
# 根据Y,计算低维的联合概率 q_ij
def q_tsne(Y):
N = np.shape(Y)[0]
sum_Y = np.sum(np.square(Y), 1)
num = -2. * np.dot(Y, Y.T)
num = 1. / (1. + np.add(np.add(num, sum_Y).T, sum_Y))
num[range(N), range(N)] = 0.
Q = num / np.sum(num)
Q = np.maximum(Q, 1e-12)
return Q, num
# datas 输入高维数据 [N,D]
# labs 高维数据的标签[N,1]
# dim 降维的维度 d
# plot 绘图
def estimate_tsen(datas, labs, dim, target_perplexity, plot=False):
N, D = np.shape(datas)
# 随机初始化低维数据Y
Y = np.random.randn(N, dim)
# 计算高维数据的联合概率
print("计算高维数据的联合概率")
P = p_joint(datas, target_perplexity)
# 开始若干轮对 P 进行放大
P = P * 4.
P = np.maximum(P, 1e-12)
# 开始进行迭代训练
# 训练相关参数
max_iter = 1500
initial_momentum = 0.5 # 摩擦系数
final_momentum = 0.8
eta = 500 # 学习率
min_gain = 0.01
dY = np.zeros([N, dim]) # 梯度
iY = np.zeros([N, dim]) # Y的变化
gains = np.ones([N, dim])
for m_iter in range(max_iter):
# 计算 Q
Q, num = q_tsne(Y)
# 计算梯度
PQ = P - Q
for i in range(N):
dY[i, :] = np.sum(np.tile(PQ[:, i] * num[:, i], (dim, 1)).T * (Y[i, :] - Y), 0)
# 带有冲量的梯度下降
if m_iter < 20:
momentum = initial_momentum
else:
momentum = final_momentum
# 如果梯度方向和增益方向相同,减小增加,反之增加
gains = (gains + 0.2) * ((dY > 0.) != (iY > 0.)) + \
(gains * 0.8) * ((dY > 0.) == (iY > 0.))
gains[gains < min_gain] = min_gain
# 当前梯度记录了上一个梯度方向信息,
iY = momentum * iY - eta * (gains * dY)
Y = Y + iY
# Y 取中心化,减均值
Y = Y - np.tile(np.mean(Y, 0), (N, 1))
# Compute current value of cost function
if (m_iter + 1) % 10 == 0:
C = np.sum(P * np.log(P / Q))
print("Iteration %d: loss is %f" % (m_iter + 1, C))
# 停止放大P
if m_iter == 100:
P = P / 4.
if plot and m_iter % 100 == 0:
print("Draw Map")
# draw_pic(Y, labs, name="%d.jpg" % (m_iter))
return Y
# 画出每次迭代的降维效果图
def draw_pic(datas, labs, name='1.jpg'):
plt.cla()
unque_labs = np.unique(labs)
colors = [plt.cm.Spectral(each)
for each in np.linspace(0, 1, len(unque_labs))]
p = []
legends = []
for i in range(len(unque_labs)):
index = np.where(labs == unque_labs[i])
pi = plt.scatter(datas[index, 0], datas[index, 1], c=[colors[i]])
p.append(pi)
legends.append(unque_labs[i])
plt.legend(p, legends)
plt.show()
if __name__ == "__main__":
data = pd.read_csv('dataset/darmanis.csv')
data = data.to_numpy().T
matrix = data[1:, 2:].astype(float)
print(matrix.shape)
labels = data[1:, 1:2]
print(labels.shape)
print(labels)
pca_matrix = pca(matrix, 30)
X = pca_matrix.real
Y = estimate_tsen(X, labels, 2, 30, plot=True)
# 降维后用kmeans聚类
centroids, clusterAssment=KMeans(Y,9)
defalut_color = ['#FF0000', '#FF1493', '#9400D3', '#7B68EE', '#FFD700',
'#00BFFF', '#00FF00', '#FF8C00', '#FF4500', '#8B4513',
'#1F77B4', '#FF7F0E', '#2CA02C', '#D62728']
# 根据不同的簇上色
for i in range(Y.shape[0]):
colorIndex = int(clusterAssment[i, 0])
plt.scatter(Y[i, 0], Y[i, 1], color=defalut_color[colorIndex])
# 绘制质心
mark = ['Dr', 'Db', 'Dg', 'Dk', '^b', '+b', 'sb', 'db', ', 'pb']
for i in range(centroids.shape[0]):
plt.plot(centroids[i, 0], centroids[i, 1], mark[i])
plt.show()
# draw_pic(Y, labels)
和PCA进行结合,先用PCA降至30维,再用TSNE降至两维以可视化展示
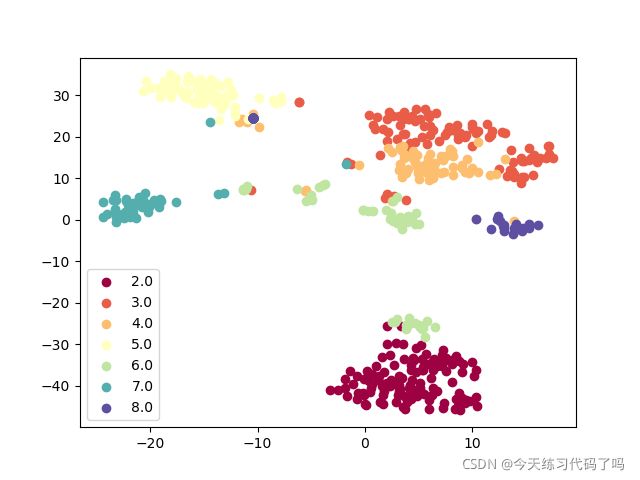
聚类效果

3.MDS
基本思想:MDS与PCA一样,是一种有效的降维方式,其可获得样本间相似性的空间表达。MDS的原理可以简述为,利用样本的成对相似性,构建一个低维空间,使每对样本在高维空间的距离与在构建的低维空间中的样本相似性尽可能保持一致。

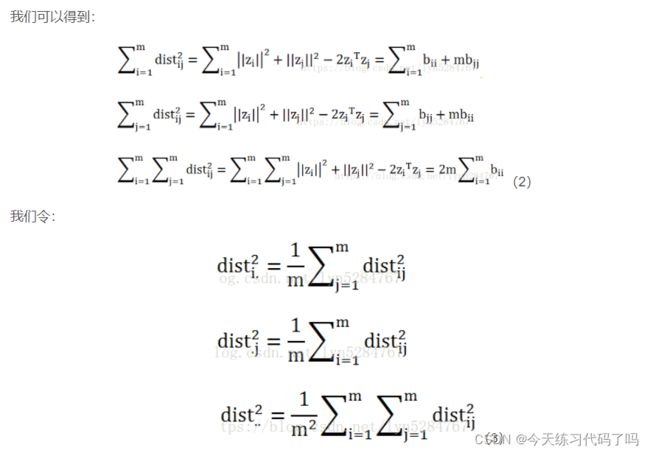
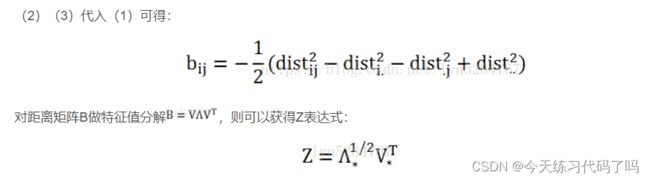
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
from K_Means import KMeans
from sklearn.manifold import TSNE
'''
MDS算法
基本思想:将高维坐标中的点投影到低维空间中,保持点彼此之间的相似性尽可能不变。
算法流程:
1.利用给定数据计算距离矩阵
2.计算降维后矢量Z的互相关矩阵B
3.对B进行特征值分解,选取较大的若干特征值与特征向量
'''
# loading dataset
def loadData(filename):
data = pd.read_csv(filename).to_numpy()
data = data.T
return data
# normalization
def normalization(data):
maxcols = data.max(axis=0)
mincols = data.min(axis=0)
data_shape = data.shape
data_rows = data_shape[0]
data_cols = data_shape[1]
nor_data = np.empty((data_rows, data_cols))
for i in range(data_cols):
nor_data[:, i] = (data[:, i] - mincols[i]) / (maxcols[i] - mincols[i])
return nor_data
# Calculate the distance between the two sample points
def calDistance(data):
n_cells, n_genes = data.shape
distance = np.zeros((n_cells, n_cells))
for i in range(n_cells):
for j in range(n_cells):
distance[i, j] = np.sqrt(np.dot((data[i] - data[j]), (data[i] - data[j]).T))
return distance
'''
parameter:
distance:The distance matrix obtained by the calDistance function
dim:The number of dimensions you want to reduce
return:Compute the cross-correlation matrix B of the dimension reduction vector Z
'''
def mds(distance, dim):
n_cells = distance.shape[0]
distance[distance < 0] = 0
distance = distance ** 2
T1 = np.ones((n_cells, n_cells)) * np.sum(distance) / n_cells ** 2
T2 = np.sum(distance, axis=1, keepdims=True) / n_cells
T3 = np.sum(distance, axis=0, keepdims=True) / n_cells
B = -(T1 - T2 - T3 + distance)
eigval, eigvec = np.linalg.eig(B)
index = np.argsort(-eigval)[:dim]
sort_eigval = eigval[index].real
sort_eigvec = eigvec[:, index]
return sort_eigvec * sort_eigval ** 0.5
# Dimensionality reduction visualization
def showDimension(data, labels):
plt.cla()
unique_labels = np.unique(labels)
colors = [plt.cm.Spectral(each) for each in np.linspace(0, 1, len(unique_labels))]
p = []
legends = []
for i in range(len(unique_labels)):
index = np.where(labels == unique_labels[i])
pi = plt.scatter(data[index, 0], data[index, 1], c=[colors[i]])
p.append(pi)
legends.append(unique_labels[i])
plt.legend(p, legends)
plt.show()
# Cluster visualization
def showCluster(dataSet, k, clusterAssment):
m, n = dataSet.shape
if n != 2:
print("Data is not two-dimensional")
return 1
defalut_color = ['#95d0fc', '#96f97b', '#c79fef',
'#ff81c0','#00035b', '#06c2ac',
'#ffff14', '#033500', '#c20078']
if k > len(defalut_color):
print("K is too big")
return 1
# Draw all samples
for i in range(m):
colorIndex = int(clusterAssment[i, 0])
plt.scatter(dataSet[i, 0], dataSet[i, 1], color=defalut_color[colorIndex])
if __name__ == '__main__':
dataset = loadData('dataset/darmanis.csv')
data = dataset[1:, 2:].astype(np.float32)
data = normalization(data)
labels = dataset[1:, 1].astype(int).flatten()
# Using MDS
distance = calDistance(data)
mds_data = mds(distance, 2)
_, clusterAssment1 = KMeans(mds_data, 9)
plt.subplot(1, 2, 1)
plt.title('MDS+K-means')
showCluster(mds_data, 9, clusterAssment1)
# showDimension(dim_data, labels)
# Using TSNE
tsne_data = TSNE(n_components=2).fit_transform(data)
_, clusterAssment2 = KMeans(tsne_data, 9)
plt.subplot(1, 2, 2)
plt.title('TSNE+K-means')
showCluster(tsne_data, 9, clusterAssment2)
plt.show()
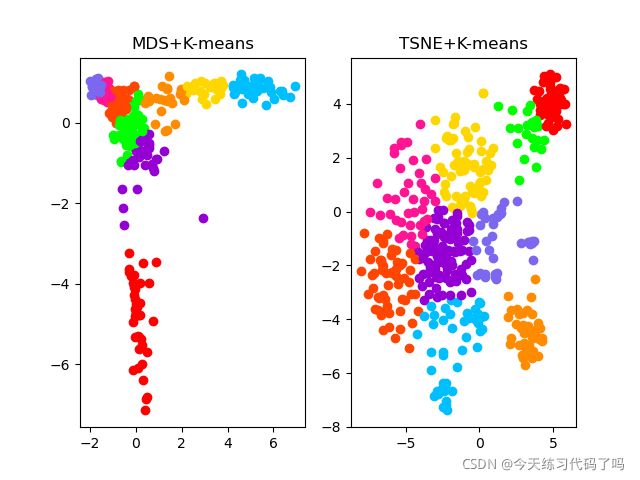
darmanis数据集聚类效果
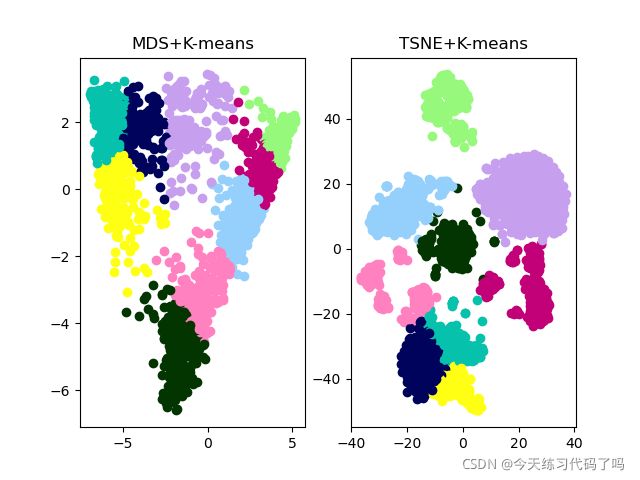
zeisel数据集聚类效果(zeisel数据集包括3005个细胞2000个高表达量基因,样本太大,我的小破电脑起码跑了五分钟才出效果)
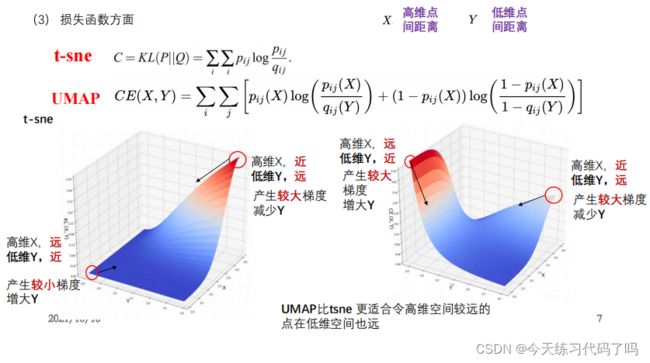
4.UMAP
UMAP与TSNE的比较:
1.比tsne可以得到更好的数据聚拢效果,能够表达更好的局部结构
2.运算效率以及运算速度比tsne好得多,可以适用于大规模数据降维
3.可以实现任意维度的降维
UMAP具体实现过程和TSNE算法类似:
1.设计一个函数(概率)来构建高位样本点,两两之间的关系(联合概率)
2.构建另一个函数(概率)来构建低维样本点两两之间的关系
3.构造一个损失函数,通过学习的方法(梯度下 降)令高维样本点之间的关系和低维样本点之间的关系尽可能相似
5.自编码器
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
import tensorflow.compat.v1 as tf
from tensorflow import keras
import os
from sklearn.cluster import KMeans
from sklearn.metrics import normalized_mutual_info_score
import umap
tf.disable_v2_behavior()
'''
C=Fc(Fm(Fa(X)))
Fc:聚类算法 Fm:流形学习器 Fa:自编码器 X:原始数据
先用自编码器降至几十维,再用umap降至两维,最后使用kmeans算法聚类
'''
os.environ["KMP_DUPLICATE_LIB_OK"] = "TRUE"
print(tf.__version__)
# 数据标准化
def processed(array):
maxcols = array.max(axis=0)
mincols = array.min(axis=0)
data_shape = array.shape
data_rows = data_shape[0]
data_cols = data_shape[1]
t = np.empty((data_rows, data_cols))
for i in range(data_cols):
t[:, i] = (array[:, i] - mincols[i]) / (maxcols[i] - mincols[i])
return t
# 聚类可视化
def draw_cluster(dataset, lab, colors):
plt.cla()
vals_lab = set(lab.tolist())
for i, val in enumerate(vals_lab):
index = np.where(lab == val)[0]
sub_dataset = dataset[index, :]
plt.scatter(sub_dataset[:, 0], sub_dataset[:, 1], s=16., color=colors[i])
dataset = pd.read_csv('dataset/darmanis.csv')
dataset = dataset.to_numpy().T
# 数据预处理,归一化,变浮点
data = dataset[1:, 2:].astype(np.float32)
data = processed(data)
labels = np.array(dataset[1:, 1:2]).flatten()
x_train = data.reshape(-1, 2000)
print(x_train.shape)
# 编码层输出维数
encoding_dim = 64
# encoder layers
input_img = keras.layers.Input(shape=(2000,))
encoded = keras.layers.Dense(512, activation='relu')(input_img)
encoded = keras.layers.Dense(256, activation='relu')(encoded)
encoded = keras.layers.Dense(128, activation='relu')(encoded)
encoder_output = keras.layers.Dense(encoding_dim)(encoded)
# decoder layers
decoded = keras.layers.Dense(128, activation='relu')(encoder_output)
decoded = keras.layers.Dense(256, activation='relu')(decoded)
decoded = keras.layers.Dense(512, activation='relu')(decoded)
decoder_output = keras.layers.Dense(2000, activation='tanh')(decoded)
# construct the autoencoder model
encoder = keras.Model(inputs=input_img, outputs=encoder_output)
autoencoder = keras.Model(inputs=input_img, outputs=decoder_output)
lr = 0.001
autoencoder.compile(optimizer=tf.train.AdamOptimizer(lr),
loss='mse',
metrics=['accuracy'])
print(autoencoder.summary())
autoencoder.fit(x_train, x_train, batch_size=512, epochs=100)
encoded_data = encoder.predict(data)
# plotting
# AE+UMAP
umap_ae_data = umap.UMAP(n_components=2, n_neighbors=30, min_dist=0.01).fit_transform(encoded_data)
km1 = KMeans(n_clusters=9).fit(umap_ae_data)
km1_labs = km1.labels_.astype(int).flatten()
print(normalized_mutual_info_score(km1_labs, labels)) # 0.7524384782147444
# UMAP
umap_data = umap.UMAP(n_components=2, n_neighbors=30, min_dist=0.01).fit_transform(data)
km2 = KMeans(n_clusters=9).fit(umap_data)
km2_labs = km2.labels_.astype(int).flatten()
print(normalized_mutual_info_score(km2_labs, labels)) # 0.6077615952027848
colors = ['#95d0fc', '#96f97b', '#c79fef',
'#ff81c0', '#00035b', '#06c2ac',
'#ffff14', '#033500', '#c20078']
plt.subplot(1, 2, 1)
draw_cluster(umap_ae_data, km1_labs, colors)
plt.subplot(1, 2, 2)
draw_cluster(umap_data, km2_labs, colors)
plt.show()