2022数维杯国际数学建模C题代码+思路+报告
2022数维杯国际数学建模C题
目录
2022数维杯国际数学建模C题
数据说明
Task1
数据分析
Task2
筛选出脑结构特征和行为结构特征
两表合并
随机森林分类
随机森林参数输出
Task3
读入数据并将ecog列删除
两表合并并编码
聚类成AD,CN,MCI三大类
从MCI中亚聚类成三个子类(SMC、EMCI、LMCI)
Task4
导入原始数据
对原始数据进行预处理
对剩余的特征进行可视化
分开CN、MCI、Dementia三个表格进行分析
Task5
解释一下:
数据说明
- COLPROT 数据收集研究协议
- ORIGPROT 原始研究方案
- PTID 兴趣倾向
- SITE 网站
- VISCODE 个人认为是根据不同的时间所运行的诊断代码
- EXAMDATE 测试日期
- DX_bl 诊断
- AGE 年龄
以上的均是每个人的基本信息,并不能称作是我们的特征(除了年龄和DX DX是我们的标签)
参与者 PT
- PT GENDER 参与者性别
- PT EDUCAT 参与者教育
- PT ETHCAT 参与者是哪里人
- PT RACCAT 参与者肤色
- PT MARRY 参与者婚姻状态
这些是可以当作我们的特征的,属于每个人的基本情况
- APOE4 治疗阿尔茨海默病(AD)的新靶点研究人员解开了APOE4基因,这是阿尔茨海默病最重要的遗传风险因素_澎湃号·湃客_澎湃新闻-The Paper
- FDG 角扣带、颞扣带和后扣带的平均FDG-PET
- PIB 额叶皮层、前扣带回、楔前叶皮层和顶叶皮层的平均PIB SUVR
- AV45 参考区域-整个小脑的氟倍他吡平均值。Freesurfer定义的区域;有关详细信息,请参阅LONI上的Jagust实验室PDFFBB 萘酚洋红
- ABETA 阿尔茨海默病重要研究因素——Aβ(β-淀粉样蛋白) - 知乎 (zhihu.com)
- TAU Tau蛋白在阿尔兹海默症中的作用| Abcam中文官网
- PTAU 阿尔兹海默症诊断重要生物标志物 ——Aβ/T-Tau/P-Tau
- CDR-SB 具有心理测量特性,使其成为全面评估AD患者认知和功能障碍的主要结局指标。它可能被证明对AD早期痴呆前期阶段的研究特别有用。
- ADAS ADAS-Cog阿尔兹海默病评定量表_上海瑞狮生物科技有限公司——康复医疗器械 (ruishihealthcare.com)
- MMSE MMSE量表_百度百科 (baidu.com)
分数在27-30分:正常
分数<27分:认知功能障碍
21-26,轻度
10-20,中度
0-9,重度
- RAVLT 精神认知表
- RAVLT_immediate 5次试验总和
- RAVLT_learning 试验5-试验1
- RAVLT_forgetting 健忘 RAVLT遗忘(试验5-延迟)
- RAVLT_perc_forgetting perc(RAVLT-遗忘百分比)
- LDELTTOTAL(逻辑记忆-延迟回忆)
- DIGITSCOR 数字符号替代
- TRAPSCOR 促甲状腺素受体抗体
- FAQ(功能活动问卷)
- MOCA 蒙特利尔认知评估量表
- ECOG评分标准_百度百科 (baidu.com)
- FLDSTRENG 磁共振视野强度
- FSVERSION Freesurfer是用于MRI图像处理
- mPACCdigit ADNI修饰的临床前阿尔茨海默氏症认知复合物(PACC),带数字符号替换
- mPACCtrailsB(ADNI修饰的临床前阿尔茨海默氏症认知复合物(PACC),带有轨迹B)
- TRABSCOR(轨迹B)
以上这些,个人认为不用纠结具体含义是什么,只需要知道它们是通过某些技术测出来的数据,就是我们要用来建立模型的关键特征
Task1
对附件数据的特征指标进行预处理,研究数据特征与阿尔茨海默病诊断的相关性。
数据分析
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
%matplotlib inline
import missingno
df = pd.read_csv('../datasets/ADNIMERGE_New.csv')
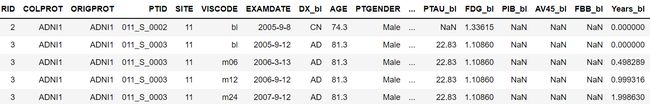
df.head()先展示下数据
我们可以发现:
- PTID的左边三个数字与SITE相同,右边的数字与RID相同,中间用‘_s_’隔开
- 而ADNI1那俩列通过查资料,是发现只是代表使用了哪份协议,和我们任务并不相关。
- VISCODE中,bl大家都知道是baseline,我个人认为是所对应的代码程序。然后经过分析,发现它与后面的M这一列数值是一致的。然后也是经过分析,它与EXAMDATE也是有关联的,具体关联是(我举个例子)
当2005-9-12号患者第一次去检验时,VISCODE标注的是bl,M标注的是0。当2006-3-13号患者第二次去检验时,VISCODE标注的是m06,M标注的是6. 我们可以观察发现,这俩日期刚好相差6个月。经过多次验证,所以我们可以推断出。bl所对的日期是患者第一次参加检验的日期,这时M是0,。mxx所对应的日期是患者经过x个月后非首次参加检验的日期,这时M是x。说白了M那一列就是患者相差几个月检验的意思。VISCODE就是根据患者的情况选择不一样的代码程序去检验。
- RID是有重复的,重复的RID均代表一个人,重复的次数说明这个人一共检验了多少次。
- 相同的RID所对应的特征_bl列的数值是相同的。但所对应的非bl后缀的特征的数值是不相同的
- 各特征都是对称的,均有含bl后缀与不含bl后缀的列。 唯独years_dl这一列是没有years列所对应的。
- DX:总共有三个类别(CN、MCI、Dementia),DX_bl总共有五个类别(CN、SMC、EMCI、LMCI、AD)
从表格暂时分析出的就这么多
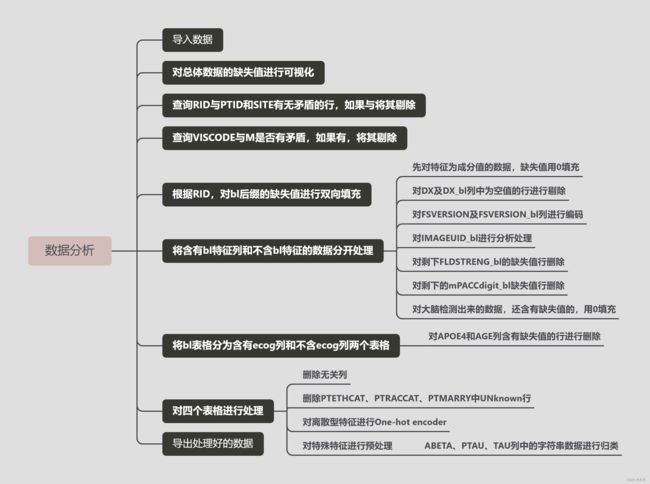
通过上面的数据分析,数据处理的思维导图在脑海里已经出来了,下面我展示一下:
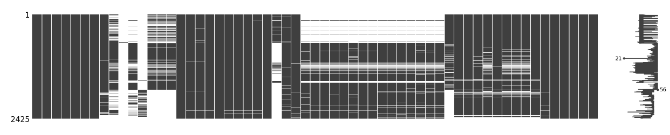
对总体数据的缺失值进行可视化
#全部数据总体缺失值可视化
missingno.matrix(df,figsize=(30,5))- 可以发现,缺失值超级多,分布都不是很良好
查看一下原始的维度
df.shape查询RID与PTID和SITE有无矛盾的行,如果与将其剔除
df['RID1'] = df['PTID'].apply(lambda x: int(x.split('_S_')[-1]))
df = df.reset_index(drop = True)
a=[]
for i in range(0,len(list(df['RID']))):
if df['RID'][i] != df['RID1'][i]:
print(df['RID'][i])
else:
a.append('%d无异常情况'%i)
len(a)--> 16222
df['SITE1'] = df['PTID'].apply(lambda x: int(x.split('_S_')[0]))
b=[]
for i in range(0,len(list(df['SITE']))):
if df['SITE'][i] != df['SITE1'][i]:
print(df['SITE'][i])
else:
b.append('%d无异常情况'%i)
len(b)--> 16222
- 可以发现,并无异常情况
查询VISCODE与M是否有矛盾,如果有,将其剔除
df.loc[df['VISCODE']=='bl','VISCODE'] = '0'
df['VISCODE'] = df['VISCODE'].apply(lambda x: int(x.split('m')[-1]) )
c=[]
for i in range(0,len(list(df['VISCODE']))):
if df['VISCODE'][i] != df['M'][i]:
print(df['VISCODE'][i])
else:
c.append('%d无异常情况'%i)
len(c)--> 16222
- 可以发现,并无异常情况
根据上文的数据分析,才会出现以上两步的异常值查找。
根据RID,对缺失值进行填充
df = pd.read_csv('../datasets/ADNIMERGE_New.csv')
rid = list(set(list(df['RID'])))
#因为发现只有后缀为bl的才有此规律,因此我们只对bl列做处理
for i in rid:
df[df['RID']==i][['IMAGEUID_bl','Ventricles_bl','Hippocampus_bl','WholeBrain_bl','Entorhinal_bl',
'Fusiform_bl','MidTemp_bl','ICV_bl','ABETA_bl','TAU_bl','PTAU_bl','FDG_bl','mPACCdigit_bl','mPACCtrailsB_bl',
'TRABSCOR_bl','DIGITSCOR_bl','LDELTOTAL_BL','RAVLT_perc_forgetting_bl','RAVLT_forgetting_bl','RAVLT_learning_bl',
'RAVLT_immediate_bl','MMSE_bl','ADASQ4_bl','ADAS13_bl','ADAS11_bl','CDRSB_bl','EXAMDATE_bl','DX_bl','AGE',
'PTGENDER','PTEDUCAT','PTETHCAT','PTRACCAT','PTMARRY','APOE4']].fillna(method='ffill',inplace=True)
df[df['RID']==i][['IMAGEUID_bl','Ventricles_bl','Hippocampus_bl','WholeBrain_bl','Entorhinal_bl',
'Fusiform_bl','MidTemp_bl','ICV_bl','ABETA_bl','TAU_bl','PTAU_bl','FDG_bl','mPACCdigit_bl','mPACCtrailsB_bl',
'TRABSCOR_bl','DIGITSCOR_bl','LDELTOTAL_BL','RAVLT_perc_forgetting_bl','RAVLT_forgetting_bl','RAVLT_learning_bl',
'RAVLT_immediate_bl','MMSE_bl','ADASQ4_bl','ADAS13_bl','ADAS11_bl','CDRSB_bl','EXAMDATE_bl','DX_bl','AGE',
'PTGENDER','PTEDUCAT','PTETHCAT','PTRACCAT','PTMARRY','APOE4']].fillna(method='bfill',inplace=True)
df = df.drop('RID',axis=1)- 填充规则是:双向填充。 因为每个RID所对应的含bl后缀的特征列的值是相同的。
将含有bl特征列和不含bl特征的数据分开处理
df = df[df['VISCODE']=='bl']先抽出所有VISCODE=bl的行,为什么呢。因为相同RID所对应的bl后缀的特征值是一样的,所以为了减少训练成本,防止过拟合现象,我们就只提取VISCODE中的bl数据进行训练。因此我选择用bl行以及bl列构成一个新的表格。
c= []
for i in list(df.columns):
if i.split('_')[-1] == 'bl' or i.split('_')[-1] == 'BL' :
c.append(i)
df_or = df.drop(c,axis=1)
b = ['PTGENDER','PTEDUCAT','PTETHCAT','PTRACCAT','PTMARRY','APOE4','M','update_stamp','AGE']
for i in b:
c.append(i)
df_bl = df[c]
df_bl.shape,df_or.shape
-->((2425, 60), (2425, 59))
- b列表里的特征是共有特征,就是个人的基本信息
- c列表里的特征是所有含bl后缀的特征
- 切分出来可以看下,含bl的表格 维度是2425,60。比不含的表格维度多一列,因为Years这特征只有bl的,没有不含bl的。
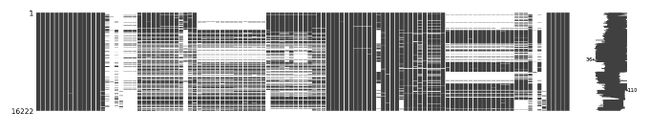
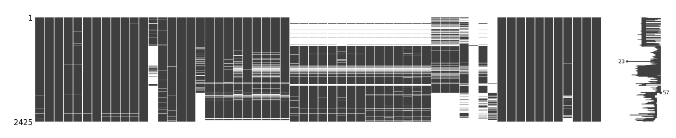
两个表格分别进行缺失值可视化
含bl:
不含bl:
对两个表格进行处理
先对特征为成分值的数据,缺失值用0填充
hanliang = ['FDG_bl','PIB_bl','AV45_bl','FBB_bl','ABETA_bl','TAU_bl','PTAU_bl','CDRSB_bl','ADAS11_bl','ADAS13_bl','ADASQ4_bl','MMSE_bl','LDELTOTAL_BL','DIGITSCOR_bl','TRABSCOR_bl','FAQ_bl','MOCA_bl','RAVLT_immediate_bl','RAVLT_learning_bl','RAVLT_forgetting_bl','RAVLT_perc_forgetting_bl']
df_bl[hanliang] = df_bl[hanliang].fillna(0)
df_or[hanliang] = df_or[hanliang].fillna(0)因为成分值的数据,如果缺失,我个人认为就是没检测到,就是没有这个含量,所以用0填充
对DX及DX_bl列中为空值的行进行剔除
df_bl = df_bl.drop(df_bl[df_bl['DX_bl'].isna()].index,axis=0)因为这是有监督的任务,标签如果是空的话那就是无监督任务了,任务无法进行,不符合题意。
对FSVERSION及FSVERSION_bl列进行编码
df_bl.loc[df_bl['FSVERSION_bl']=='Cross-Sectional FreeSurfer (FreeSurfer Version 4.3)','FLDSTRENG_bl'] = 1
df_bl.loc[df_bl['FSVERSION_bl']=='Cross-Sectional FreeSurfer (5.1)','FLDSTRENG_bl'] = 2
df_bl.loc[df_bl['FSVERSION_bl']=='Cross-Sectional FreeSurfer (6.0)','FLDSTRENG_bl'] = 3
df_or.loc[df_or['FSVERSION']=='Cross-Sectional FreeSurfer (FreeSurfer Version 4.3)','FLDSTRENG_bl'] = 1
df_or.loc[df_or['FSVERSION']=='Cross-Sectional FreeSurfer (5.1)','FLDSTRENG_bl'] = 2
df_or.loc[df_or['FSVERSION']=='Cross-Sectional FreeSurfer (6.0)','FLDSTRENG_bl'] = 3
为什么进行label encoder呢,因为机器不认识字符串,想利用这特征只能转换成数字,而且这特征是有层次关系的,因此不选用One-hot encoder
对IMAGEUID_bl进行分析处理
df1 = df[df['FLDSTRENG_bl']=='1.5 Tesla MRI']
#发现只有一条数据是大于20W的,因此我们将它剔除,并拿20W当作阈值
df1 = df1.drop(df1[df1['IMAGEUID_bl']> 200000].index,axis=0)
df_bl = df_bl.drop(df1[df1['IMAGEUID_bl']> 200000].index,axis=0)
df2 = df[df['FLDSTRENG_bl']=='3 Tesla MRI']
df3 = df_bl[df_bl['FLDSTRENG_bl']==3]
#取20W和80W当成阈值
df_bl.loc[(df_bl['IMAGEUID_bl']> 200000) & (df_bl['IMAGEUID_bl']< 800000) ,'FLDSTRENG_bl'] = 2
df_bl.loc[df_bl['IMAGEUID_bl']< 200000,'FLDSTRENG_bl'] = 1
df_bl.loc[df_bl['IMAGEUID_bl']> 800000,'FLDSTRENG_bl'] = 3不含bl的代码不粘贴了,与上方一致的
- 通过分析,IMAGEUID与FLDSTRENG是有关系的。
- 共分为三个区间,IMAGEUID<200000的数据,对应的FLDSTRENG的空值(这里我们编码为1)
- IMAGEUID>200000的数据,对应的FLDSTRENG的1.5 Tesla MRI(这里我们编码为2)
- IMAGEUID>800000的数据,对应的FLDSTRENG的3 Tesla MRI(这里我们编码为3)
对剩下FLDSTRENG_bl的缺失值行删除
#将剩下FLDSTRENG_bl的缺失值行删除,因为剩下的行中,FLDSTRENG_bl FSVERSION_bl IMAGEUID_bl同时缺失
df_bl = df_bl.drop(df_bl[df_bl['FLDSTRENG_bl'].isna()].index,axis=0)
对剩下的mPACCdigit_bl缺失值行删除
#将剩下的mPACCdigit_bl缺失值行删除,因为剩下的两行中,mPACCdigit_bl mPACCtrailsB_bl同时缺失
df_bl = df_bl.drop(df_bl[df_bl['mPACCdigit_bl'].isna()].index,axis=0)对大脑检测出来的数据,还含有缺失值的,用0填充
#用0填充
a = ['Ventricles_bl','Hippocampus_bl','WholeBrain_bl','Entorhinal_bl','Fusiform_bl','MidTemp_bl','ICV_bl']
df_bl[a] = df_bl[a].fillna(0)
因为我个人认为,筛选到这一步,还有缺失值的,也只能是未检测到该成分才会缺失,因此我拿0填充
将bl表格分为含有ecog列和不含ecog列两个表格
ecog = ['EcogPtMem_bl','EcogPtLang_bl','EcogPtVisspat_bl','EcogPtPlan_bl','EcogPtOrgan_bl','EcogPtDivatt_bl','EcogPtTotal_bl',
'EcogSPMem_bl','EcogSPLang_bl','EcogSPVisspat_bl','EcogSPPlan_bl','EcogSPOrgan_bl','EcogSPDivatt_bl','EcogSPTotal_bl']
df_bl_notec = df_bl.drop(ecog,axis=1)
# 筛选出所有不含缺失值的行
df_bl_ec = df_bl[df_bl[ecog].isna().T.any() == False]- 因为ecog代表的是随访记录,也是缺失值含有最多的特征列。
- 因为我个人认为这些没数据的,有可能是前面的数据体现它并不具有继续被研究的价值
- 或者就是失访(由于事故无法确定该人情况)
对APOE4和AGE列含有缺失值的行进行删除
df_bl_notec = df_bl_notec.drop(df_bl_notec[df_bl_notec['APOE4'].isna()].index,axis=0)
df_bl_ec = df_bl_ec.drop(df_bl_ec[df_bl_ec['APOE4'].isna()].index,axis=0)
df_bl_notec = df_bl_notec.drop(df_bl_notec[df_bl_notec['AGE'].isna()].index,axis=0)
df_bl_ec = df_bl_ec.drop(df_bl_ec[df_bl_ec['AGE'].isna()].index,axis=0)
- APOE4是代表新靶点,AGE是年龄,这两个都无法用普通的插值方法处理,因为0,1,2,3....都具有不同含义。而且APOE4是离散型,所以不能用均值填补等方法,因此选择直接剔除。
处理到此,所有缺失值和异常值都处理了,得到了四份完美的数据。(一份bl有ecog,一份bl无ecog,一份无bl有ecog,一份无bl无ecog)
可视化处理完的结果:
有ecog的两表情况
不含ecog的两表情况
写出第一次处理完的表格
df_or_notec.to_csv('../mid_data/df_or_notec.csv',index=False)
df_or_ec.to_csv('../mid_data/df_or_ec.csv',index=False)
df_bl_notec.to_csv('../mid_data/df_bl_notec.csv',index=False)
df_bl_ec.to_csv('../mid_data/df_bl_ec.csv',index=False)继续对那四个表格进行处理
DX_map = {
'CN': 1,
'SMC': 2,
'EMCI': 3,
'LMCI': 4,
'AD': 5,
}
F_map = {
'Cross-Sectional FreeSurfer (5.1)':2,
'Cross-Sectional FreeSurfer (FreeSurfer Version 4.3)':1,
'Cross-Sectional FreeSurfer (6.0)':3
}
df_bl_ec['DX_bl'] = df_bl_ec['DX_bl'].map(DX_map)
df_bl_ec['FSVERSION_bl'] = df_bl_ec['FSVERSION_bl'].map(F_map)- 对特征DX进行label encoder,以方便后面的相关性分析和建模
- 还有其它字符串类型的特征进行label encoder
删除无关列
df_bl_ec = df_bl_ec.drop(['EXAMDATE_bl','Years_bl','Month_bl','M','update_stamp'],axis=1)
删除UNknown行
df_bl_ec = df_bl_ec.drop(df_bl_ec[df_bl_ec['PTETHCAT'] =='Unknown'].index,axis=0)
df_bl_ec = df_bl_ec.drop(df_bl_ec[df_bl_ec['PTRACCAT'] =='Unknown'].index,axis=0)
df_bl_ec = df_bl_ec.drop(df_bl_ec[df_bl_ec['PTMARRY'] =='Unknown'].index,axis=0)- 经过数据分析,以上三个特征是离散型特征,且含有Unknown数据的,因此我们对含有Unknown的进行剔除
对离散型特征进行One-hot encoder
one_hot_df = pd.get_dummies(df_bl_ec['PTETHCAT'])
df_bl_ec = pd.merge(df_bl_ec,one_hot_df,left_index=True,right_index=True)
df_bl_ec = df_bl_ec.drop('PTETHCAT',axis=1)
one_hot_df = pd.get_dummies(df_bl_ec['PTRACCAT'])
df_bl_ec = pd.merge(df_bl_ec,one_hot_df,left_index=True,right_index=True)
df_bl_ec = df_bl_ec.drop('PTRACCAT',axis=1)
one_hot_df = pd.get_dummies(df_bl_ec['PTMARRY'])
df_bl_ec = pd.merge(df_bl_ec,one_hot_df,left_index=True,right_index=True)
df_bl_ec = df_bl_ec.drop('PTMARRY',axis=1)
one_hot_df = pd.get_dummies(df_bl_ec['PTGENDER'])
df_bl_ec = pd.merge(df_bl_ec,one_hot_df,left_index=True,right_index=True)
df_bl_ec = df_bl_ec.drop('PTGENDER',axis=1)对特殊特征进行预处理
df_bl_ec.loc[df_bl_ec['ABETA_bl']=='>1700','ABETA_bl'] = 1701
df_bl_ec.loc[df_bl_ec['PTAU_bl']=='<8','PTAU_bl'] = 7
df_bl_ec.loc[df_bl_ec['ABETA_bl']=='<200','ABETA_bl'] = 199
df_bl_ec.loc[df_bl_ec['PTAU_bl']=='>120','PTAU_bl'] = 121
df_bl_ec.loc[df_bl_ec['TAU_bl']=='>1300','TAU_bl'] = 1301
df_bl_ec.loc[df_bl_ec['TAU_bl']=='<80','TAU_bl'] = 79
df_bl_ec.loc[df_bl_ec['ABETA_bl'].astype(float)>1700,'ABETA_bl'] = 1701
df_bl_ec.loc[df_bl_ec['PTAU_bl'].astype(float)<8,'PTAU_bl'] = 7
df_bl_ec.loc[df_bl_ec['ABETA_bl'].astype(float)<200,'ABETA_bl'] = 199
df_bl_ec.loc[df_bl_ec['PTAU_bl'].astype(float)>120,'PTAU_bl'] = 121
df_bl_ec.loc[df_bl_ec['TAU_bl'].astype(float)>1300,'TAU_bl'] = 1301
df_bl_ec.loc[df_bl_ec['TAU_bl'].astype(float)<80,'TAU_bl'] = 79- 通过数据分析发现,以上这几列特征,在表格中除了有数值型特征外还含有字符串型特征,因此我们要对字符串类型进行数值编码
- 例如ABETA 这一列,它里面含有很多不一样的数值,但也有“>1700”这样子的字符串,我猜这数据是想让我们将>1700数据的值按一个类别的方式进行处理,<1700的数值按连续型特征处理
- 因此我们将所有类别根据要求,都取一个固定的数去代表这个类别。例如>1700我们取1701
- 继续通过分析发现同一列在不同的表格里,居然会多出几个不一样的类别,例如“>1700”"<200",为了数据的统一,例如将不同表格中<200的数都换成同一数值代替。这样子对四个表格同时处理。
到此,第二次处理完,输出数据
df_or_notec.to_csv('../mid_data/df_or_notec_encoding.csv',index=False)
df_or_ec.to_csv('../mid_data/df_or_ec_encoding.csv',index=False)
df_bl_notec.to_csv('../mid_data/df_bl_notec_encoding.csv',index=False)
df_bl_ec.to_csv('../mid_data/df_bl_ec_encoding.csv',index=False)
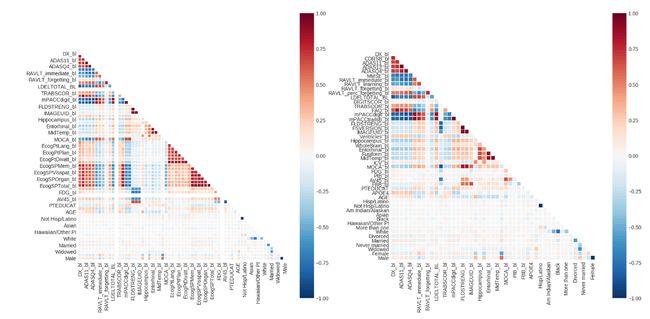
对经过两次处理的含有bl的表格进行相关性分析
corr()
分别绘制两个热力图,左图是有ecog特征的,右图是没有ecog特征的
# 绘制两个数据集的热力图
plt.style.use('seaborn-whitegrid') # 设置绘图风格
fig = plt.figure(figsize=(20,10))
# 绘制第一个热力图
plt.subplot(1,2,1)
mask = np.zeros_like(df_bl_ec.corr(),dtype=np.bool)
mask[np.triu_indices_from(mask)] = True
sns.heatmap(df_bl_ec.corr(),
vmin=-1,vmax=1,
square=True,
cmap=sns.color_palette("RdBu_r",100),
mask=mask,
linewidths=.5)
# 绘制第二个热力图
plt.subplot(1,2,2)
mask = np.zeros_like(df_bl_notec.corr(),dtype=np.bool)
mask[np.triu_indices_from(mask)] = True
sns.heatmap(df_bl_notec.corr(),
vmin=-1,vmax=1,
square=True,
cmap=sns.color_palette("RdBu_r",100),
mask=mask,
linewidths=.5)- 论文里也要分开提到有ecog和无ecog相关性的区别。
- 从作图可以看出,有ecog的特征,相关性还挺高的
- 这个是用了corr()默认的相关性检验方法,好像是皮尔逊Pearson相关系数
随机森林特征重要性查看
第一题还没完,单独一个corr()并不能解决问题。我选择用随机森林去拟合数据,从而查看特征的重要性
#df_bl_ec
from sklearn.ensemble import RandomForestClassifier
from sklearn import metrics
from sklearn.metrics import r2_score #用于模型拟合优度评估
label=df_bl_ec['DX_bl']
train=df_bl_ec.drop(['DX_bl'],axis=1)
#构造随机森林模型
model=RandomForestClassifier()
model.fit(train,label) #模型拟合
predictions= model.predict(train) #预测
print("train r2:%.3f"%r2_score(label,model.predict(train))) #评估--> r2:1.00
可以发现拟合得非常完美,因此我们就不对随机森林调优了
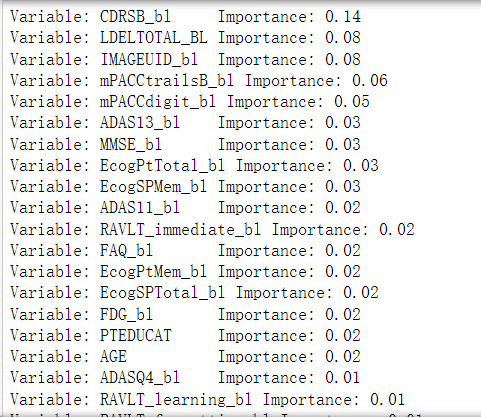
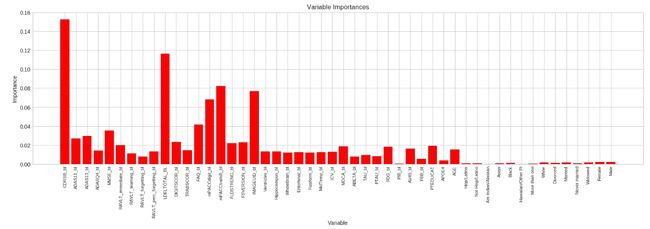
直接查看特征重要性
feature_list = list(train.columns)
importances = list(model.feature_importances_)
feature_importances = [(feature, round(importance, 2)) for feature, importance in zip(feature_list,importances)] #将相关变量名称与重要性对应
feature_importances = sorted(feature_importances, key = lambda x: x[1], reverse = True) #排序
[print('Variable: {:12} Importance: {}'.format(*pair)) for pair in feature_importances] #输出特征影响程度详细数据
以上只是其中一个表格的重要性,另一个看代码吧。对俩表特征重要性进行可视化
含ecog的:
不含ecog的
Task2
利用所附的脑结构特征和认知行为特征,设计阿尔茨海默病的智能诊断。
筛选出脑结构特征和行为结构特征
df_bl_ec = pd.read_csv('../mid_data/df_bl_ec_encoding.csv')
df_bl_notec = pd.read_csv('../mid_data/df_bl_notec_encoding.csv')
#筛选出脑结构特征和行为结构特征
f_col = ['IMAGEUID_bl','Ventricles_bl','Hippocampus_bl','WholeBrain_bl','Entorhinal_bl','Fusiform_bl','MidTemp_bl','ICV_bl',
'RAVLT_immediate_bl','RAVLT_learning_bl','RAVLT_forgetting_bl','RAVLT_perc_forgetting_bl','LDELTOTAL_BL','DX_bl',
'MOCA_bl','mPACCdigit_bl','mPACCtrailsB_bl']
df_bl_ec = df_bl_ec[f_col]
df_bl_notec = df_bl_notec[f_col] - 读入刚刚处理完的两个表格
- f_col列表里面的特征就是筛选出来的脑结构特征和行为结构特征。筛选方法就是,如果这个特征属于脑结构或行为结构就直接拿过来
- 然后分别放入含ecog和不含ecog的表格中
两表合并
#因为脑结构特征和行为结构特征都没有ecog,所以俩表拼接
df_bl = df_bl_ec.append(df_bl_notec)
df_bl = df_bl.reset_index(drop=True)
df_bl.to_csv('../mid_data/df_bl_merge.csv')- 因为脑结构特征和行为结构特征都没有ecog,ecog代表的是随访的记录。因此直接将两表拼接,都去除ecog特征,然后将拼接后的表格输出
随机森林分类
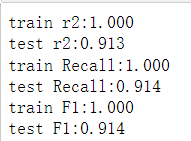
因为本赛题是多分类,这里是五分类问题。由于本赛题的数据属于结构化数据,因此我们选择了树模型,不选择深度学习。评价指标这里我们用了,r2,recall,F1,论文里要对这三个指标进行介绍
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import train_test_split
from sklearn import metrics
from sklearn.metrics import r2_score #用于模型拟合优度评估
label=df_bl['DX_bl']
train=df_bl.drop(['DX_bl'],axis=1)
train_x,test_x,train_y,test_y=train_test_split(train,label,test_size=0.3,random_state=4396)
model=RandomForestClassifier()
model.fit(train_x,train_y)
predictions= model.predict(test_x)
print("train r2:%.3f"%r2_score(train_y,model.predict(train_x)))
print("test r2:%.3f"%r2_score(test_y,predictions))
print("train Recall:%.3f"%metrics.recall_score(train_y,model.predict(train_x),average='micro'))
print("test Recall:%.3f"%metrics.recall_score(test_y,predictions,average='micro'))
print("train F1:%.3f"%metrics.f1_score(train_y,model.predict(train_x),average='macro'))
print("test F1:%.3f"%metrics.f1_score(test_y,predictions,average='macro'))
- 可以发现效果也是极其不错的,训练集的拟合都是完全正确的,验证集都打到了0.9以上
- 在本次随机森林中,因为数据量不大,所以n_estimators我们使用默认的100,max_depth让它扩展直到所有叶子是纯的
随机森林参数输出
model.estimators_
model.estimators_[0].random_state--> 466110319
Task3
首先,将CN、MCI和AD聚类为三大类。然后,针对MCI中包含的三个子类(SMC、EMCI和LMCI),继续将聚类细化为三个子类。
读入数据并将ecog列删除
df_bl_ec = pd.read_csv('../mid_data/df_bl_ec_encoding.csv')
df_bl_notec = pd.read_csv('../mid_data/df_bl_notec_encoding.csv')
#为了利用到更多的特征,在聚类前,将ecog的删除,然后两表合并以换取更多特征
df_bl_ec = df_bl_ec.drop(['EcogPtMem_bl',
'EcogPtLang_bl', 'EcogPtVisspat_bl', 'EcogPtPlan_bl', 'EcogPtOrgan_bl',
'EcogPtDivatt_bl', 'EcogPtTotal_bl', 'EcogSPMem_bl', 'EcogSPLang_bl',
'EcogSPVisspat_bl', 'EcogSPPlan_bl', 'EcogSPOrgan_bl',
'EcogSPDivatt_bl', 'EcogSPTotal_bl'],axis=1)- 因为ecog列包含太多缺失值,我们聚类也不可能分为两个表格聚类了,不然的话会造成很大的数据损失,也会影响聚类效果
两表合并并编码
df = df_bl_ec.append(df_bl_notec)
df = df.reset_index(drop=True)
df.loc[df['DX_bl']==1,'DX_three'] = 1
df.loc[df['DX_bl']==2,'DX_three'] = 2
df.loc[df['DX_bl']==3,'DX_three'] = 2
df.loc[df['DX_bl']==4,'DX_three'] = 2
df.loc[df['DX_bl']==5,'DX_three'] = 3
df.loc[df['DX_bl']==2,'DX_son'] = 2
df.loc[df['DX_bl']==3,'DX_son'] = 3
df.loc[df['DX_bl']==4,'DX_son'] = 4- 注意:我们将AD,CN归为1和5。属于MCI的类都归为2
- 然后再MCI类里,MCI中的三个子类分别编码为2,3,4
聚类成AD,CN,MCI三大类
这里我们使用了KMeans聚类算法
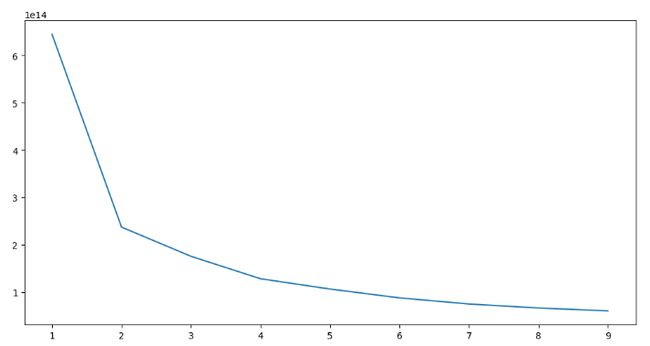
首先寻找最优K值
from sklearn.cluster import KMeans
## 寻找最佳K值
x = df[df.columns.difference(['DX_bl','DX_three','DX_son'])]
inertia = []
for i in range(1,10):
km = KMeans(n_clusters=i,n_jobs=-1)
km.fit(x)
inertia.append(km.inertia_) #簇内的误差平方和
plt.figure(figsize=(12,6))
plt.plot(range(1,10),inertia)
plt.show()
##手肘原则- 根据手肘原则,最优K值是2,我尝试过删除特征使K等于三,但聚类出来的效果太过于平均,与原标签分布情况不符。
- 因此我想在不删除标签的情况下,先进行二聚类,再从二聚类里面选一个标签,再对它进行二聚类,这样就完成了聚成三大类的目的
第一次聚类
我们选k = 2进行第一次聚类
km = KMeans(n_clusters=2,n_jobs=-1)
y_means = km.fit_predict(x)
df['Kmeans_three'] = y_means
df.loc[df['Kmeans_three']==1,'Kmeans_three'] = 2
df_1 = df[df['Kmeans_three']==0]
- 上图是第一次聚类的标签分布图
- 根绝原始标签的分布,我们选择对0这一类进行二次聚类
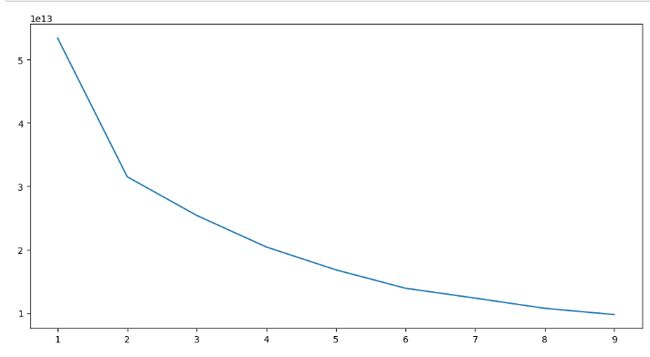
第二次寻找最优K值
- 它依旧是2,因此说明0这一类,让它再聚成两类是合理的,也验证了我们思路的正常
第二次聚类
km = KMeans(n_clusters=2,n_jobs=-1)
y_means = km.fit_predict(x)
df_1['Kmeans_three'] = y_means
df_1.loc[df_1['Kmeans_three']==0,'Kmeans_three'] = 3
df['Kmeans_three'][df_1['Kmeans_three'].index] = df_1['Kmeans_three']
df['Kmeans_three'].value_counts()
这是聚成AD、CN、MCI三类的分布情况
从MCI中亚聚类成三个子类(SMC、EMCI、LMCI)
将MCI抽出来,并寻找最佳K值
df_MCI = df[df['Kmeans_three']==2]
## 寻找最佳K值
x = df_MCI[df_MCI.columns.difference(['DX_bl','DX_three','DX_son'])]
inertia = []
for i in range(1,10):
km = KMeans(n_clusters=i,n_jobs=-1)
km.fit(x)
inertia.append(km.inertia_) #簇内的误差平方和
plt.figure(figsize=(12,6))
plt.plot(range(1,10),inertia)
plt.show()
- 方法和原因和上面聚类的一样
- 这里我们也是分为两次聚类
- 也是因为它的最佳K值等于2
聚类完的标签分布:
Task4
附件中的同一样本包含不同时间点的特征,请将其与时间点的关系进行分析,以揭示其中的模式,不同种类疾病随时间的演变。
导入原始数据
df = pd.read_csv('../datasets/ADNIMERGE_New.csv')对原始数据进行预处理
df = df.drop(['COLPROT','ORIGPROT','PTID','SITE','VISCODE'],axis=1)
c= []
for i in list(df.columns):
if i.split('_')[-1] == 'bl' or i.split('_')[-1] == 'BL' :
c.append(i)
b = ['PTGENDER','PTEDUCAT','PTETHCAT','PTRACCAT','PTMARRY','APOE4','update_stamp','AGE']
for i in b:
c.append(i)
df_or = df.drop(c,axis=1)
# 对特征为成分值的数据,缺失值用0填充
hanliang = ['FDG','PIB','AV45','FBB','ABETA',
'TAU','PTAU','CDRSB','ADAS11','ADAS13','ADASQ4',
'MMSE','LDELTOTAL','DIGITSCOR','TRABSCOR','FAQ',
'MOCA','RAVLT_immediate','RAVLT_learning',
'RAVLT_forgetting','RAVLT_perc_forgetting']
df_or[hanliang] = df_or[hanliang].fillna(0)
df_or = df_or.drop(df_or[df_or['DX'].isna()].index,axis=0)
df_or.loc[df_or['FSVERSION']=='Cross-Sectional FreeSurfer (FreeSurfer Version 4.3)','FLDSTRENG_bl'] = 1
df_or.loc[df_or['FSVERSION']=='Cross-Sectional FreeSurfer (5.1)','FLDSTRENG_bl'] = 2
df_or.loc[df_or['FSVERSION']=='Cross-Sectional FreeSurfer (6.0)','FLDSTRENG_bl'] = 3
df1 = df[df['FLDSTRENG']=='1.5 Tesla MRI']
df2 = df[df['FLDSTRENG']=='3 Tesla MRI']
df_or = df_or.drop(df1[df1['IMAGEUID']> 200000].index,axis=0)
#取20W和80W当成阈值
df_or.loc[(df_or['IMAGEUID']> 200000) & (df_or['IMAGEUID']< 800000) ,
'FLDSTRENG'] = 2
df_or.loc[df_or['IMAGEUID']< 200000,'FLDSTRENG'] = 1
df_or.loc[df_or['IMAGEUID']> 800000,'FLDSTRENG'] = 3
df_or = df_or.drop(df_or[df_or['FLDSTRENG'].isna()].index,axis=0)
df_or = df_or.drop(df_or[df_or['mPACCdigit'].isna()].index,axis=0)
#用0填充
a = ['Ventricles','Hippocampus','WholeBrain','Entorhinal','Fusiform',
'MidTemp','ICV']
df_or[a] = df_or[a].fillna(0)
ecog = ['EcogPtMem','EcogPtLang','EcogPtVisspat','EcogPtPlan','EcogPtOrgan',
'EcogPtDivatt','EcogPtTotal',
'EcogSPMem','EcogSPLang','EcogSPVisspat','EcogSPPlan','EcogSPOrgan',
'EcogSPDivatt','EcogSPTotal']
df_or_notec = df_or.drop(ecog,axis=1)
df_or_notec.loc[df_or_notec['ABETA']=='>1700','ABETA'] = 1701
df_or_notec.loc[df_or_notec['ABETA']=='<200','ABETA'] = 199
df_or_notec.loc[df_or_notec['PTAU']=='<8','PTAU'] = 7
df_or_notec.loc[df_or_notec['PTAU']=='>120','PTAU'] = 121
df_or_notec.loc[df_or_notec['TAU']=='>1300','TAU'] = 1301
df_or_notec.loc[df_or_notec['TAU']=='<80','TAU'] = 79
df_or_notec.loc[df_or_notec['ABETA'].astype(float)>1700,'ABETA'] = 1701
df_or_notec.loc[df_or_notec['PTAU'].astype(float)<8,'PTAU'] = 7
df_or_notec.loc[df_or_notec['ABETA'].astype(float)<200,'ABETA'] = 199
df_or_notec.loc[df_or_notec['PTAU'].astype(float)>120,'PTAU'] = 121
df_or_notec.loc[df_or_notec['TAU'].astype(float)>1300,'TAU'] = 1301
df_or_notec.loc[df_or_notec['TAU'].astype(float)<80,'TAU'] = 79
df_or_notec = df_or_notec.drop(['Month','EXAMDATE'],axis=1)
df_or_notec = df_or_notec.reset_index(drop = True)- 上方的预处理与任务一的预处理一模一样
- 删除无关列,删除含bl的列
- 对特征为成分值的数据,缺失值用0填充
- 对FSVERSION进行label encoder
- 对DX含有缺失值的行进行剔除
- 根据IMAGEUID的三个阈值,对FLDSTRENG进行编码
- 删除FLDSTRENG和mPACCdigit还含有缺失值的行,原因是与它们相关的那几列在这几行同时含有缺失值,无法进行推断填充,因此删除
- 因为ecog属于随访的特征,与本任务无关,因此删除
- 对ABETA,PTAU这两列中的字符串特征进行label encoder 连续型特征维持原状
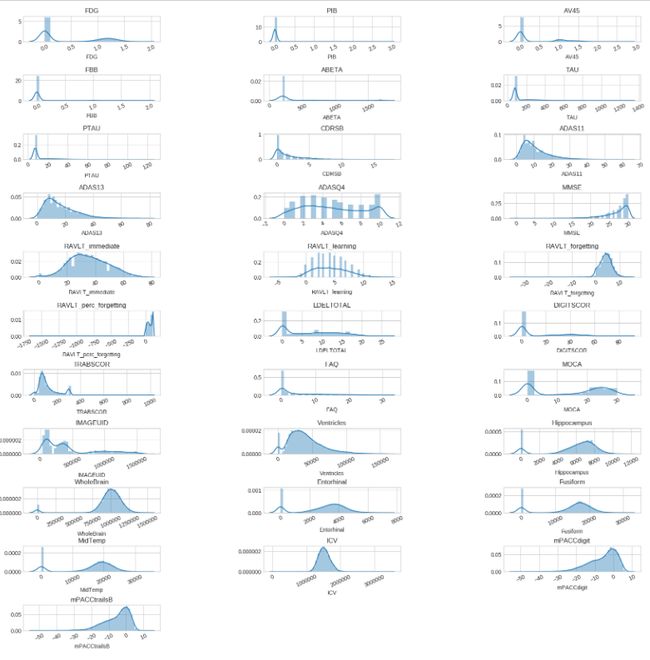
对剩余的特征进行可视化
col= ['FDG', 'PIB', 'AV45', 'FBB', 'ABETA', 'TAU', 'PTAU', 'CDRSB',
'ADAS11', 'ADAS13', 'ADASQ4', 'MMSE', 'RAVLT_immediate',
'RAVLT_learning', 'RAVLT_forgetting', 'RAVLT_perc_forgetting',
'LDELTOTAL', 'DIGITSCOR', 'TRABSCOR', 'FAQ', 'MOCA', 'IMAGEUID', 'Ventricles', 'Hippocampus', 'WholeBrain',
'Entorhinal', 'Fusiform', 'MidTemp', 'ICV', 'mPACCdigit',
'mPACCtrailsB']
import math
def plot_distribution(dataset,cols=5,width=20,height=15,hspace=0.2,wspace=0.5):
plt.style.use('seaborn-whitegrid')
fig = plt.figure(figsize=(width,height))
fig.subplots_adjust(left=None,bottom=None,right=None,top=None,wspace=wspace,hspace=hspace)
rows = math.ceil(float(dataset.shape[1]) / cols)
#以上都是准备工作
for i,column in enumerate(dataset.columns):
ax = fig.add_subplot(rows,cols,i+1)
ax.set_title(column)
g = sns.distplot(dataset[column]) #绘制分布图
plt.xticks(rotation=25)- 选出剩余的特征,并封装一个绘图函数
- 我们选择对每个特征进行绘制概率密度图,以便筛选特征
df_or_notec_draw = df_or_notec[col]
import warnings
warnings.filterwarnings('ignore')
plot_distribution(df_or_notec_draw,cols=3,width=20,height=20,hspace=1.3,wspace=0.75)
- 从中可以一眼看出,哪些特征是离散型特征,哪些特征是连续型特征,哪些特征对我们的任务讨论有意义
- 在这里,我们筛选掉所有离散型特征,和所有基本是0的特征
- 因为本任务让我们研究不同种类的疾病与时间的关系,离散型特征是类别,不能放映出其关系,全是0的数据更没研究价值
col1 = ['mPACCtrailsB','mPACCdigit','ICV','MidTemp','Fusiform',
'Entorhinal','WholeBrain','Hippocampus','Ventricles','IMAGEUID',
'TRABSCOR','RAVLT_forgetting','RAVLT_immediate','ADAS13','ADAS11']
def plot(dataset,cols=5,width=20,height=15,hspace=0.2,wspace=0.5):
plt.style.use('seaborn-whitegrid')
fig = plt.figure(figsize=(width,height))
fig.subplots_adjust(left=None,bottom=None,right=None,top=None,wspace=wspace,hspace=hspace)
rows = math.ceil(float(dataset.shape[1]) / cols)
#以上都是准备工作
for i,column in enumerate(dataset.columns):
ax = fig.add_subplot(rows,cols,i+1)
ax.set_title(column)
g = sns.lineplot(dataset[column].index,dataset[column])
plt.xticks(rotation=25)- coll是筛选后的特征
- 再定义一个绘图函数,里面是根据筛选出来的特征后画折线图,以便反映该特征随时间的变化的关系
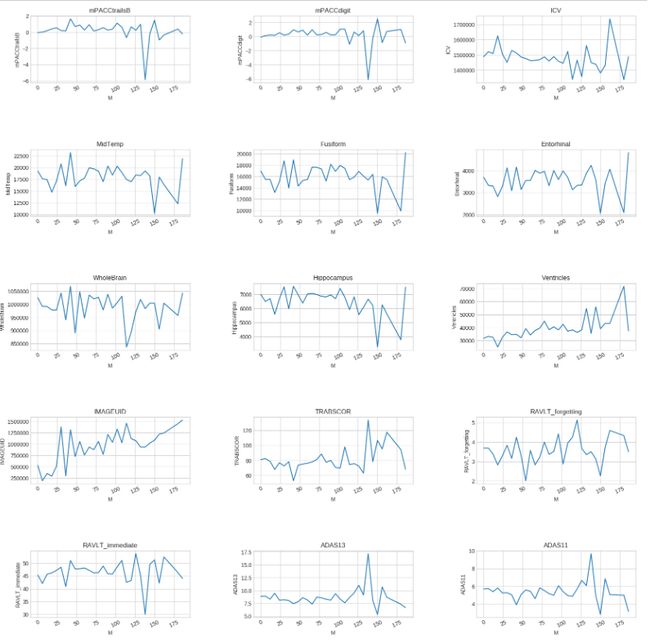
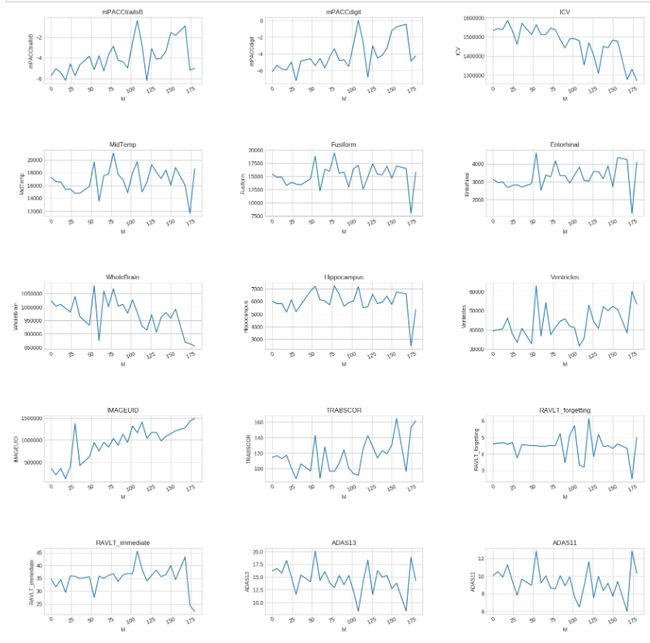
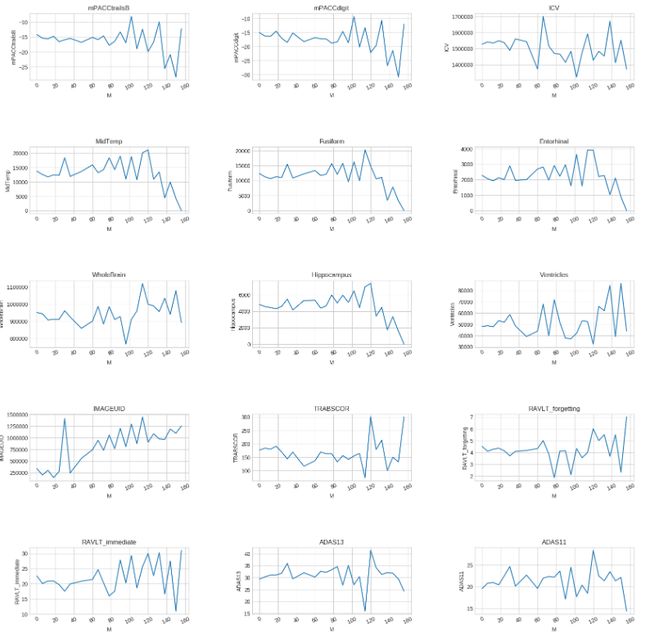
分开CN、MCI、Dementia三个表格进行分析
df_CN = df_or_notec[df_or_notec['DX']=='CN']
df_MCI = df_or_notec[df_or_notec['DX']=='MCI']
df_DE = df_or_notec[df_or_notec['DX']=='Dementia']CN
df_CN_draw = df_CN.groupby('M')[col1].mean()
plot(df_CN_draw,cols=3,width=20,height=20,hspace=1,wspace=0.4)
MCI
df_MCI_draw = df_MCI.groupby('M')[col1].mean()
plot(df_MCI_draw,cols=3,width=20,height=20,hspace=1,wspace=0.4)
Dementia
df_DE_draw = df_DE.groupby('M')[col1].mean()
plot(df_DE_draw,cols=3,width=20,height=20,hspace=1,wspace=0.4)
- 以上是三个不同的疾病中的特征随着时间变化的关系
- 分析一下,在论文里吹一下
Task5
CN、SMC、EMCI、LMCI、AD五类的早期干预及诊断标准请查阅相关文献。
解释一下:
为什么要提取VISCODE中的bl数据,因为相同RID所对应的bl后缀的特征值是一样的,所以为了减少训练成本,防止过拟合现象,我们就只提取VISCODE中的bl数据进行训练。
这次分享主要目的是交流学习,这也仅代表我一人的解题思路,不一定正确,也请各位大佬多多指正!