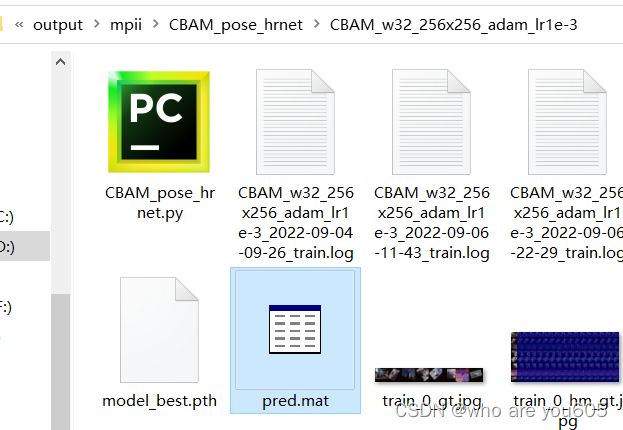
MPII单人人体姿态估计数据集测试结果(test)
一、MPII数据集下载
http://human-pose.mpi-inf.mpg.de/#download
在这下载的Annotations标注文件是matlab格式

所以根据HRNet源代码来下载注释文件

二、在MPII数据集上测试test
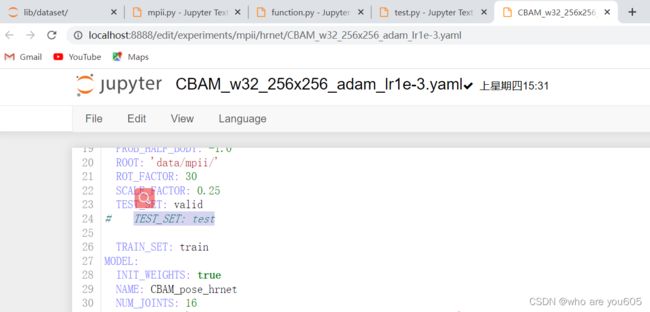
1.将配置文件中的TEST_SET由*valid转变为test*
读取test.json文件
2.获取测试结果pred.mat
在虚拟环境中运行以下命令行:
python tools/test.py --cfg experiments/mpii/hrnet/CBAM_w32_256x256_adam_lr1e-3.yaml TEST.MODEL_FILE output/mpii/CBAM_pose_hrnet/CBAM_w32_256x256_adam_lr1e-3/model_best.pth
三、matlab获取测试结果
1.在MPII官网下载评估工具包

2.解压evalMPII.zip得到eval文件夹
根据github提供方法进行matlab操作
注意:eval文件夹中还需包含一个ground_truth文件夹,其中包含两个文件:
annolist_dataset_v12.mat和mpii_human_pose_v1_u12.mat
这两个文件本来可以在MPII官网上下载得到,但现在没有资源了,可以从一个github网站上获取:Github3G/prepare-and-test-on-MPII
其中的eval_test文件夹中就包含所需文件。

3.将eval文件夹中的evaluatePCKh.m使用matlab打开,并修改为如下代码:
(运行原来的evaluatePCKh.m会报错:
evaluatePCKh()
输入参数的数目不足。
出错 evaluatePCKh (line 15)
tableTex = cell(length(predidxs)+1,1);
)
所以修改为:
% Evaluate performance by comparing predictions to ground truth annotations.
%%% OPTIONS %%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
% IDs of prediction sets to include in results
PRED_IDS = [1, 2, 3, 4, 5, 6];
% Subset of the data that the predictions correspond to ('val' or 'train')
plotcurve = false;
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
addpath ('eval')
fprintf('# MPII single-person pose evaluation script\n')
range = 0:0.01:0.5;
tableDir = './latex'; if (~exist(tableDir,'dir')), mkdir(tableDir); end
plotsDir = './plots'; if (~exist(plotsDir,'dir')), mkdir(plotsDir); end
tableTex = cell(length(PRED_IDS)+1,1);
% load ground truth
p = getExpParams(-1)
load([p.gtDir '/annolist_dataset_v12'], 'annolist');
load([p.gtDir '/mpii_human_pose_v1_u12'], 'RELEASE');
annolist_test = annolist(RELEASE.img_train == 0);
% evaluate on the "single person" subset only
single_person_test = RELEASE.single_person(RELEASE.img_train == 0);
% convert to annotation list with a single pose per entry
[annolist_test_flat, single_person_test_flat] = flatten_annolist(annolist_test,single_person_test);
% represent ground truth as a matrix 2x14xN_images
gt = annolist2matrix(annolist_test_flat(single_person_test_flat == 1));
% compute head size
headSize = getHeadSizeAll(annolist_test_flat(single_person_test_flat == 1));
pckAll = zeros(length(range),16,length(PRED_IDS));
for i = 1:length(PRED_IDS);
% load predictions
p = getExpParams(PRED_IDS(i));
try
load(p.predFilename, 'preds');
catch
preds = h5read(p.predFilename, '/preds');
end
if size(preds, 1) == 2
preds = permute(preds, [3, 2, 1]);
end
% Check that there are the same number of predictions and ground truth
% annotations. If this assertion fails, a likely cause is a mismatch in
% subsets (eg predictions are for the training set but ground truth
% annotations are for the validation set).
fprintf('%d\n', length(preds))
fprintf('%d\n', length(gt))
assert(length(preds) == length(gt));
pred_flat = annolist_test_flat(single_person_test_flat == 1);
for idx = 1:length(preds);
for pidx = 1:length(pred_flat(idx).annorect.annopoints.point);
joint = pred_flat(idx).annorect.annopoints.point(pidx).id + 1;
xy = preds(idx, joint, :);
pred_flat(idx).annorect.annopoints.point(pidx).x = xy(1);
pred_flat(idx).annorect.annopoints.point(pidx).y = xy(2);
end
end
% pred = annolist2matrix(pred_flat(single_person_flat == 1));
pred = annolist2matrix(pred_flat);
% only gt is allowed to have NaN
pred(isnan(pred)) = inf;
% compute distance to ground truth joints
dist = getDistPCKh(pred,gt,headSize);
% compute PCKh
pck = computePCK(dist,range);
% plot results
[row, header] = genTablePCK(pck(end,:),p.name);
tableTex{1} = header;
tableTex{i+1} = row;
pckAll(:,:,i) = pck;
auc = area_under_curve(scale01(range),pck(:,end));
fprintf('%s, AUC: %1.1f\n',p.name,auc);
end
% Save results
fid = fopen([tableDir '/pckh.tex'],'wt');assert(fid ~= -1);
for i=1:length(tableTex),fprintf(fid,'%s\n',tableTex{i}); end; fclose(fid);
% plot curves
bSave = true;
if (plotcurve)
plotCurveNew(squeeze(pckAll(:,end,:)),range,PRED_IDS,'PCKh total, MPII',[plotsDir '/pckh-total-mpii'],bSave,range(1:5:end));
plotCurveNew(squeeze(mean(pckAll(:,[1 6],:),2)),range,PRED_IDS,'PCKh ankle, MPII',[plotsDir '/pckh-ankle-mpii'],bSave,range(1:5:end));
plotCurveNew(squeeze(mean(pckAll(:,[2 5],:),2)),range,PRED_IDS,'PCKh knee, MPII',[plotsDir '/pckh-knee-mpii'],bSave,range(1:5:end));
plotCurveNew(squeeze(mean(pckAll(:,[3 4],:),2)),range,PRED_IDS,'PCKh hip, MPII',[plotsDir '/pckh-hip-mpii'],bSave,range(1:5:end));
plotCurveNew(squeeze(mean(pckAll(:,[7 12],:),2)),range,PRED_IDS,'PCKh wrist, MPII',[plotsDir '/pckh-wrist-mpii'],bSave,range(1:5:end));
plotCurveNew(squeeze(mean(pckAll(:,[8 11],:),2)),range,PRED_IDS,'PCKh elbow, MPII',[plotsDir '/pckh-elbow-mpii'],bSave,range(1:5:end));
plotCurveNew(squeeze(mean(pckAll(:,[9 10],:),2)),range,PRED_IDS,'PCKh shoulder, MPII',[plotsDir '/pckh-shoulder-mpii'],bSave,range(1:5:end));
plotCurveNew(squeeze(mean(pckAll(:,[13 14],:),2)),range,PRED_IDS,'PCKh head, MPII',[plotsDir '/pckh-head-mpii'],bSave,range(1:5:end));
end
display('Done.')
4.修改后运行报错:
错误使用 load
无法读取文件 './ground_truth//annolist_dataset_v12'。没有此类文件或目录。
出错 evaluatePCKh (line 24)
load([p.gtDir '/annolist_dataset_v12'], 'annolist');
将文件路径改为绝对路径即可:
p的获取调用了getExpParams.m,因此修改此文件下p.gtDir的路径。

5.继续报错:

可知需要引用的文件并不存在或路径不正确。观察输出结果:
p的输出并未包含predFilename,也就是p.predFilename并不存在。

从调用p的程序getExpParams.m可以发现程序会跳过中间部分,无法获取到p.predFilename(此时switch中的predidx取值为-1):


所以将p.predFilename定义在switch语句之外并采用绝对路径:

6.报错:

这个报错原因是采用的pred.mat文件时val数据集的结果,而不是test数据集的结果,两个.mat文件的数据图片数量分别为2958和7247,无法匹配。所以需要进行步骤二的操作。
7.报错与5.同理,处理方法也一致:
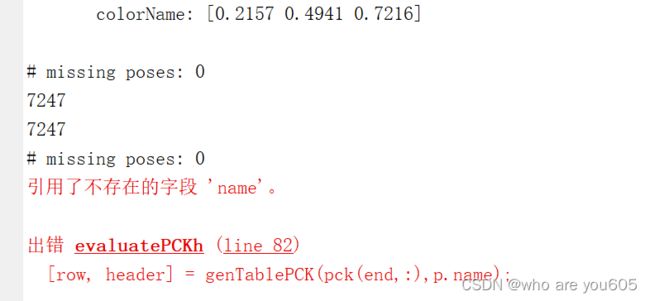

8.获得MPII数据集上的测试结果:
# MPII single-person pose evaluation script
p =
包含以下字段的 struct:
gtDir: 'D:/HRNet-Human-Pose-Estimation-master/evalMPII/eval/ground_truth/'
colorIdxs: [1 1]
partNames: {1×15 cell}
predFilename: 'D:/HRNet-Human-Pose-Estimation-master/evalMPII/eval/pred.mat'
name: 'experiment name'
colorName: [0.2157 0.4941 0.7216]
# missing poses: 0
7247
7247
# missing poses: 0
&Head & Shoulder & Elbow & Wrist & Hip & Knee & Ankle & UBody & Total \\
experiment name& 97.9 & 95.6 & 89.6 & 84.5 & 89.0 & 84.2 & 80.2 & 89.9 & 89.2 \\
experiment name, AUC: 60.0
7247
7247
# missing poses: 0
&Head & Shoulder & Elbow & Wrist & Hip & Knee & Ankle & UBody & Total \\
experiment name& 97.9 & 95.6 & 89.6 & 84.5 & 89.0 & 84.2 & 80.2 & 89.9 & 89.2 \\
experiment name, AUC: 60.0
7247
7247
# missing poses: 0
&Head & Shoulder & Elbow & Wrist & Hip & Knee & Ankle & UBody & Total \\
experiment name& 97.9 & 95.6 & 89.6 & 84.5 & 89.0 & 84.2 & 80.2 & 89.9 & 89.2 \\
experiment name, AUC: 60.0
7247
7247
# missing poses: 0
&Head & Shoulder & Elbow & Wrist & Hip & Knee & Ankle & UBody & Total \\
experiment name& 97.9 & 95.6 & 89.6 & 84.5 & 89.0 & 84.2 & 80.2 & 89.9 & 89.2 \\
experiment name, AUC: 60.0
7247
7247
# missing poses: 0
&Head & Shoulder & Elbow & Wrist & Hip & Knee & Ankle & UBody & Total \\
experiment name& 97.9 & 95.6 & 89.6 & 84.5 & 89.0 & 84.2 & 80.2 & 89.9 & 89.2 \\
experiment name, AUC: 60.0
7247
7247
# missing poses: 0
&Head & Shoulder & Elbow & Wrist & Hip & Knee & Ankle & UBody & Total \\
experiment name& 97.9 & 95.6 & 89.6 & 84.5 & 89.0 & 84.2 & 80.2 & 89.9 & 89.2 \\
experiment name, AUC: 60.0
Done.