Unet系列论文总结
Unet系类算法为什么在医疗图像处理上取得这么好的效果?很大一部分原因是由于医疗图像自身的特性所决定的,首先是医疗图像结构固定且简单、其次Unet网络结构简单与难获取的医疗图像匹配度较高。导致的问题就是深度学习强大的特征表示能力并不能很好的发挥其作用,最后就是数据集获取难度很大。一般人获取不到,就算获取到大量的医疗图像数据,对其进行标注真的是一件特别无聊的工作。专业的事交给专业的人去做比较好,现实生活中可能会存在以下几个问题,搞CV的人对医疗图像进行标注有困难,正常不正常傻傻分不清。医生对CV可能也比较陌生,不是针对所有人,针对大部分人而言来说,情况就是这么个情况。
1.Unet
U-Net: Convolutional Networks for Biomedical Image Segmentation
code
paper

从Unet网络结构图可知,该网络结构是一个编码解码结构,编码由卷积、池化组成,也可理解为下采样操作。目的是为了获取不同尺寸的feature map。解码器由卷积、特征拼接、上采样组成,其中特征拼接是feature map维度上的拼接,其目的是为了获取更厚的feature map,因编码阶段卷积池化过程会使图像的细节信息丢失,feature map进行拼接是为了尽可能的找回编码阶段所丢失的图像细节信息。拼接这一操作虽然可以避免图像信息的缺失,但这一操作能找回丢失信息的多少?拼接的方式到底好还是不好?是一个值得考虑的问题。Unet网络已然成为当前医疗图像分割的baseline,主要是因为医疗图像本身自带的一些数据特性所决定的。大多医疗图像数据的特点有以下几点:首先是医疗图像语义相比较自然场景其语义较为简单且结构固定,导致其所有的feature都很重要,也即是说低级、高级的语义信息都尽量保存下来,以便模型能更好的对其进行学习。其次是医疗数据获取难度大,能获取的数据量过少,导致的问题是网络过深与数据量少这一矛盾。会出现过拟合现象。最后是Unet相比较于其他的分割模型,其结构简单,具有更大的操作空间。
后续的改进工作大多围绕特征提取、特征拼接展开
torch实现如下
import torch
import torch.nn as nn
import torch.nn.functional as F
class double_conv2d_bn(nn.Module):
def__init__(self,in_channels,out_channels,kernel_size=3,strides=1,padding=1):
super(double_conv2d_bn,self).__init__()
self.conv1 = nn.Conv2d(in_channels,out_channels,
kernel_size=kernel_size,
stride = strides,padding=padding,bias=True)
self.conv2 = nn.Conv2d(out_channels,out_channels,
kernel_size = kernel_size,
stride = strides,padding=padding,bias=True)
self.bn1 = nn.BatchNorm2d(out_channels)
self.bn2 = nn.BatchNorm2d(out_channels)
def forward(self,x):
out = F.relu(self.bn1(self.conv1(x)))
out = F.relu(self.bn2(self.conv2(out)))
return out
class deconv2d_bn(nn.Module):
def __init__(self,in_channels,out_channels,kernel_size=2,strides=2):
super(deconv2d_bn,self).__init__()
self.conv1 = nn.ConvTranspose2d(in_channels,out_channels,
kernel_size = kernel_size,
stride = strides,bias=True)
self.bn1 = nn.BatchNorm2d(out_channels)
def forward(self,x):
out = F.relu(self.bn1(self.conv1(x)))
return out
class Unet(nn.Module):
def __init__(self):
super(Unet,self).__init__()
self.layer1_conv = double_conv2d_bn(1,8)
self.layer2_conv = double_conv2d_bn(8,16)
self.layer3_conv = double_conv2d_bn(16,32)
self.layer4_conv = double_conv2d_bn(32,64)
self.layer5_conv = double_conv2d_bn(64,128)
self.layer6_conv = double_conv2d_bn(128,64)
self.layer7_conv = double_conv2d_bn(64,32)
self.layer8_conv = double_conv2d_bn(32,16)
self.layer9_conv = double_conv2d_bn(16,8)
self.layer10_conv = nn.Conv2d(8,1,kernel_size=3,
stride=1,padding=1,bias=True)
self.deconv1 = deconv2d_bn(128,64)
self.deconv2 = deconv2d_bn(64,32)
self.deconv3 = deconv2d_bn(32,16)
self.deconv4 = deconv2d_bn(16,8)
self.sigmoid = nn.Sigmoid()
def forward(self,x):
conv1 = self.layer1_conv(x)
pool1 = F.max_pool2d(conv1,2)
conv2 = self.layer2_conv(pool1)
pool2 = F.max_pool2d(conv2,2)
conv3 = self.layer3_conv(pool2)
pool3 = F.max_pool2d(conv3,2)
conv4 = self.layer4_conv(pool3)
pool4 = F.max_pool2d(conv4,2)
conv5 = self.layer5_conv(pool4)
convt1 = self.deconv1(conv5)
concat1 = torch.cat([convt1,conv4],dim=1)
conv6 = self.layer6_conv(concat1)
convt2 = self.deconv2(conv6)
concat2 = torch.cat([convt2,conv3],dim=1)
conv7 = self.layer7_conv(concat2)
convt3 = self.deconv3(conv7)
concat3 = torch.cat([convt3,conv2],dim=1)
conv8 = self.layer8_conv(concat3)
convt4 = self.deconv4(conv8)
concat4 = torch.cat([convt4,conv1],dim=1)
conv9 = self.layer9_conv(concat4)
outp = self.layer10_conv(conv9)
outp = self.sigmoid(outp)
return outp
model = Unet()
inp = torch.rand(10,1,224,224)
outp = model(inp)
print(outp.shape)
2.DC-Unet
DC-UNet: Rethinking the U-Net Architecture with Dual Channel Efficient CNN for Medical Images Segmentation
问题:Unet方法已经成为当前主流的医学图像分割算法,然而由于原始的Unet网络主要由编码解码器构成。不能高效的提取图像特征,
解决方法:设计了高效的CNN架构以取代编码器和解码器、应用残差模块替换编码器和解码器之间的跳过连接,以改进现有的U-Net模型。
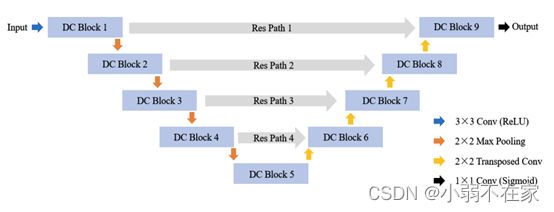
网络结构与Unet并无太大区别,
keras实现
# -*- coding: utf-8 -*-
import os
import cv2
import numpy as np
from tqdm import tqdm
import matplotlib.pyplot as plt
from keras import initializers
from keras.layers import SpatialDropout2D, Input, Conv2D, MaxPooling2D, Conv2DTranspose, concatenate, AveragePooling2D, \
UpSampling2D, BatchNormalization, Activation, add, Dropout, Permute, ZeroPadding2D, Add, Reshape
from keras.models import Model, model_from_json
from keras.optimizers import Adam
from keras.layers.advanced_activations import ELU, LeakyReLU, ReLU, PReLU
from keras.utils.vis_utils import plot_model
from keras import backend as K
from sklearn.model_selection import train_test_split
from sklearn.metrics import classification_report
from keras import applications, optimizers, callbacks
import matplotlib
import keras
import tensorflow as tf
from keras.layers import *
def conv2d_bn(x, filters, num_row, num_col, padding='same', strides=(1, 1), activation='relu', name=None):
'''
2D Convolutional layers
Arguments:
x {keras layer} -- input layer
filters {int} -- number of filters
num_row {int} -- number of rows in filters
num_col {int} -- number of columns in filters
Keyword Arguments:
padding {str} -- mode of padding (default: {'same'})
strides {tuple} -- stride of convolution operation (default: {(1, 1)})
activation {str} -- activation function (default: {'relu'})
name {str} -- name of the layer (default: {None})
Returns:
[keras layer] -- [output layer]
'''
x = Conv2D(filters, (num_row, num_col), strides=strides, padding=padding, use_bias=False)(x)
x = BatchNormalization(axis=3, scale=False)(x)
if (activation == None):
return x
x = Activation(activation, name=name)(x)
return x
def trans_conv2d_bn(x, filters, num_row, num_col, padding='same', strides=(2, 2), name=None):
'''
2D Transposed Convolutional layers
Arguments:
x {keras layer} -- input layer
filters {int} -- number of filters
num_row {int} -- number of rows in filters
num_col {int} -- number of columns in filters
Keyword Arguments:
padding {str} -- mode of padding (default: {'same'})
strides {tuple} -- stride of convolution operation (default: {(2, 2)})
name {str} -- name of the layer (default: {None})
Returns:
[keras layer] -- [output layer]
'''
x = Conv2DTranspose(filters, (num_row, num_col), strides=strides, padding=padding)(x)
x = BatchNormalization(axis=3, scale=False)(x)
return x
def DCBlock(U, inp, alpha=1.67):
'''
DC Block
Arguments:
U {int} -- Number of filters in a corrsponding UNet stage
inp {keras layer} -- input layer
Returns:
[keras layer] -- [output layer]
'''
W = alpha * U
# shortcut = inp
# shortcut = conv2d_bn(shortcut, int(W*0.167) + int(W*0.333) +
# int(W*0.5), 1, 1, activation=None, padding='same')
conv3x3_1 = conv2d_bn(inp, int(W * 0.167), 3, 3,
activation='relu', padding='same')
conv5x5_1 = conv2d_bn(conv3x3_1, int(W * 0.333), 3, 3,
activation='relu', padding='same')
conv7x7_1 = conv2d_bn(conv5x5_1, int(W * 0.5), 3, 3,
activation='relu', padding='same')
out1 = concatenate([conv3x3_1, conv5x5_1, conv7x7_1], axis=3)
out1 = BatchNormalization(axis=3)(out1)
conv3x3_2 = conv2d_bn(inp, int(W * 0.167), 3, 3,
activation='relu', padding='same')
conv5x5_2 = conv2d_bn(conv3x3_2, int(W * 0.333), 3, 3,
activation='relu', padding='same')
conv7x7_2 = conv2d_bn(conv5x5_2, int(W * 0.5), 3, 3,
activation='relu', padding='same')
out2 = concatenate([conv3x3_2, conv5x5_2, conv7x7_2], axis=3)
out2 = BatchNormalization(axis=3)(out2)
out = add([out1, out2])
out = Activation('relu')(out)
out = BatchNormalization(axis=3)(out)
return out
def ResPath(filters, length, inp):
'''
ResPath
Arguments:
filters {int} -- [description]
length {int} -- length of ResPath
inp {keras layer} -- input layer
Returns:
[keras layer] -- [output layer]
'''
shortcut = inp
shortcut = conv2d_bn(shortcut, filters, 1, 1,
activation=None, padding='same')
out = conv2d_bn(inp, filters, 3, 3, activation='relu', padding='same')
out = add([shortcut, out])
out = Activation('relu')(out)
out = BatchNormalization(axis=3)(out)
for i in range(length - 1):
shortcut = out
shortcut = conv2d_bn(shortcut, filters, 1, 1,
activation=None, padding='same')
out = conv2d_bn(out, filters, 3, 3, activation='relu', padding='same')
out = add([shortcut, out])
out = Activation('relu')(out)
out = BatchNormalization(axis=3)(out)
return out
def DCUNet(height, width, channels):
'''
DC-UNet
Arguments:
height {int} -- height of image
width {int} -- width of image
n_channels {int} -- number of channels in image
Returns:
[keras model] -- MultiResUNet model
'''
inputs = Input((height, width, channels))
dcblock1 = DCBlock(32, inputs)
pool1 = MaxPooling2D(pool_size=(2, 2))(dcblock1)
dcblock1 = ResPath(32, 4, dcblock1)
dcblock2 = DCBlock(32 * 2, pool1)
pool2 = MaxPooling2D(pool_size=(2, 2))(dcblock2)
dcblock2 = ResPath(32 * 2, 3, dcblock2)
dcblock3 = DCBlock(32 * 4, pool2)
pool3 = MaxPooling2D(pool_size=(2, 2))(dcblock3)
dcblock3 = ResPath(32 * 4, 2, dcblock3)
dcblock4 = DCBlock(32 * 8, pool3)
pool4 = MaxPooling2D(pool_size=(2, 2))(dcblock4)
dcblock4 = ResPath(32 * 8, 1, dcblock4)
dcblock5 = DCBlock(32 * 16, pool4)
up6 = concatenate([Conv2DTranspose(
32 * 8, (2, 2), strides=(2, 2), padding='same')(dcblock5), dcblock4], axis=3)
dcblock6 = DCBlock(32 * 8, up6)
up7 = concatenate([Conv2DTranspose(
32 * 4, (2, 2), strides=(2, 2), padding='same')(dcblock6), dcblock3], axis=3)
dcblock7 = DCBlock(32 * 4, up7)
up8 = concatenate([Conv2DTranspose(
32 * 2, (2, 2), strides=(2, 2), padding='same')(dcblock7), dcblock2], axis=3)
dcblock8 = DCBlock(32 * 2, up8)
up9 = concatenate([Conv2DTranspose(32, (2, 2), strides=(
2, 2), padding='same')(dcblock8), dcblock1], axis=3)
dcblock9 = DCBlock(32, up9)
conv10 = conv2d_bn(dcblock9, 1, 1, 1, activation='sigmoid')
model = Model(inputs=[inputs], outputs=[conv10])
return model
3.TransUNet: Transformers Make Strong Encoders for Medical Image Segmentation
动机:深度学习算法在医疗图像处理领域已经取得很大的成功,众多医疗场景中以医疗图像分割为例。因医疗图像本身自带的一些特性,使得Unet在医疗图像分割领域已经成为一个baseline。Unet本质上还是卷积池化构成多尺度特征,这个过程会让图像的一些细节信息丢失。也即是说提出更强的编码解码器有助于缓解图像信息丢失,于是利用transformer与Unet相结合,提出TransUnet用于医疗图像分割,并通过实验验证TransUnet的有效性。
transUnet网络结构如下图所示
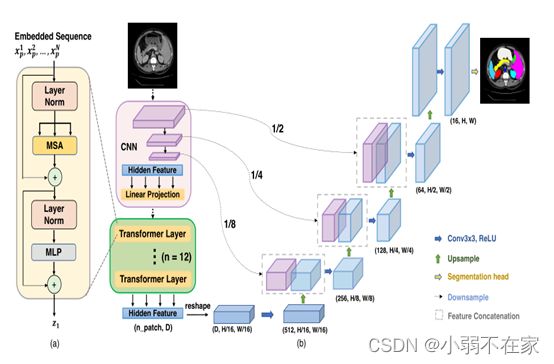
Transformer表示快被玩坏了!!!!
总结与吐槽
看了大概十篇左右Unet系列的论文,与原始Unet相比,性能虽然有一定的提升。但是模型的复杂程度也在增加,参数量、计算时间大大增多。也不是说小修小补不可以,大部分人还是做着小修小补的工作。并没有太多的亮点,哈哈哈,借用老板的话来说你就算做不出来也要学会去评判一个工作是否是个好工作。
期待看到有突破性的工作。