生信学习——GEO数据挖掘
步骤
- STEP1:表达矩阵ID转换
- STEP2:差异分析
- STEP3:KEGG数据库注释
- 完整代码
写在前面——按照生信技能树的学习路线,学完R语言就该学习GEO数据挖掘了。有人说GEO数据挖掘可以快速发文(https://zhuanlan.zhihu.com/p/36303146),不知道靠不靠谱。反正学一学总没有坏处。看完Jimmy老师的视频,写一篇总结方便日后复习。这里有很多操作在 《生信人的20个R语言习题》都可以见到,那里写的更加详细。
视频教程:https://www.bilibili.com/video/BV1is411H7Hq?p=1
代码地址:https://github.com/jmzeng1314/GEO
STEP1:表达矩阵ID转换
首先理解下面的4个概念:
GEO Platform (GPL)
GEO Sample (GSM)
GEO Series (GSE)
GEO Dataset (GDS)
理解起来也很容易。一篇文章可以有一个或者多个GSE数据集,一个GSE里面可以有一个或者多个GSM样本。多个研究的GSM样本可以根据研究目的整合为一个GDS,不过GDS本身用的很少。而每个数据集都有着自己对应的芯片平台,就是GPL。
用R获取芯片探针与基因的对应关系三部曲-bioconductor
http://www.bio-info-trainee.com/1399.html
# setwd(dir = "geo_learn/")
### step 1 ###
# 获得GSE数据集的表达矩阵
if(F){
suppressPackageStartupMessages(library(GEOquery))
gset <- getGEO('GSE42872', destdir=".",
AnnotGPL = F,
getGPL = F)
save(gset,file='GSE42872_gset.Rdata')
}
load('GSE42872_gset.Rdata')
exprSet <- exprs(gset[[1]])
pdata <- pData(gset[[1]])
group_list <- c(rep('control', 3), rep('case', 3))
# 以下操作等同于exprs(gset[[1]])
# a <- read.table("GSE42872_series_matrix.txt.gz",
# sep = "\t", quote = "", fill = T,
# comment.char = "!", header = T)
# rownames(a) <- a[,1]
# a <- a[,-1]
### step 2 ###
# 根据gset中的Annotation: GPL6244找到对应的R包,安装并使用
# BiocManager::install("hugene10sttranscriptcluster.db")
suppressPackageStartupMessages(library(hugene10sttranscriptcluster.db))
# 找不到对应R包的可以使用下面这种方法
# gpl <- getGE0('GPL6480', destdir=".")
# colnames(Table(gpl))##[1] 41108 17
# head(Table(gpL)[,c(1,6,7)])
# ## you need to check this,which column do you need
# write.csv(Table(gpl)[,c(1,6,7)],"GPL6400.csv")
### step 3 ###
# 获得探针与基因的对应关系,对表达矩阵进行ID转换
ls("package:hugene10sttranscriptcluster.db")
ids <- toTable(hugene10sttranscriptclusterSYMBOL)
# 将表达矩阵中没有对应基因名字的探针去除
table(rownames(exprSet) %in% ids$probe_id)
dim(exprSet)
exprSet <- exprSet[(rownames(exprSet) %in% ids$probe_id),]
dim(exprSet)
# 将exprSet与ids的数据顺序一一对应
ids <- ids[match(rownames(exprSet),ids$probe_id),]
dim(ids)
# 整合表达矩阵
# 多个探针对应一个基因的情况下,只保留在所有样本里面平均表达量最大的那个探针。
tmp <- by(exprSet,ids$symbol,
function(x) rownames(x)[which.max(rowMeans(x))])
tmp[1:20]
probes <- as.character(tmp)
exprSet <- exprSet[rownames(exprSet) %in% probes, ]
dim(exprSet)
dim(ids)
rownames(exprSet) <- ids[match(rownames(exprSet),ids$probe_id),2]
save(exprSet, group_list, file = 'GSE42872_new_exprSet.Rdata')
STEP2:差异分析
load('GSE42872_new_exprSet.Rdata')
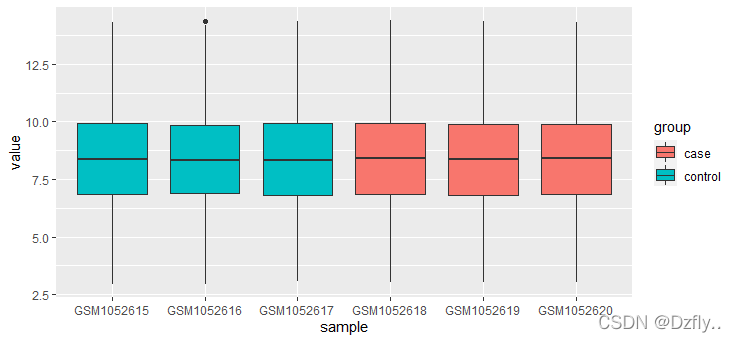
# 绘制boxplot,看数据分布是否整齐
library(reshape2)
m_exprSet <- melt(exprSet)
head(m_exprSet)
colnames(m_exprSet) <- c("symbol", "sample", "value")
head(m_exprSet)
m_exprSet$group <- rep(group_list, each = nrow(exprSet))
head(m_exprSet)
library(ggplot2)
ggplot(m_exprSet, aes(x = sample, y = value, fill = group)) + geom_boxplot()
# clustering
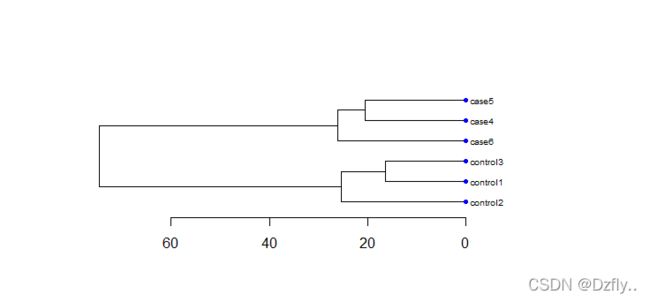
# 看聚类效果,效果好则说明数据可用
colnames(exprSet) <- paste(group_list,1:6,sep='')
hc <- hclust(dist(t(exprSet)))
nodePar <- list(lab.cex = 0.6, pch = c(NA, 19), cex = 0.7, col = "blue")
par(mar=c(5,5,5,10))
plot(as.dendrogram(hc), nodePar = nodePar, horiz = TRUE)
# 使用limma进行差异分析
library(limma)
# 得到按组分离的矩阵
design <- model.matrix(~0 + factor(group_list))
colnames(design) <- levels(factor(group_list))
rownames(design) <- colnames(exprSet)
design
# 差异比较矩阵
contrast.matrix <- makeContrasts("case-control" ,levels = design)
# contrast.matrix <- makeContrasts("control-case" ,levels = design)
contrast.matrix
##step1
# 在给定一系列序列的情况下,对每个基因拟合线性模型
# exprSet要求行对应于基因,列对应于样本
# design要求行对应样本,列对应系数
fit <- lmFit(exprSet,design)
##step2
# 根据lmFit的拟合结果进行统计推断,计算给定一组对比的估计系数和标准误差
# fit由lmFit得到的
# contrasts要求:行对应拟合系数,列包含对比度
fit2 <- contrasts.fit(fit, contrast.matrix)
# Methods of assessing differential expression
fit2 <- eBayes(fit2)
##step3
# 从线性模型拟合中提取出排名靠前的基因表
# For topTable, fit should be an object of class MArrayLM as produced by lmFit and eBayes.
# topTable 默认显示前10个基因的统计数据;使用选项n可以设置,n=Inf就是不设上限,全部输出
# 只有case-control一组的差异基因,就用coef = 1
tempOutput <- topTable(fit2, coef=1, n=Inf)
# 去除缺失值
nrDEG <- na.omit(tempOutput)
#write.csv(nrDEG2,"limma_notrend.results.csv",quote = F)
head(nrDEG)
## volcano plot
DEG <- nrDEG
# 设定阈值,选出UP、DOWN、NOT表达基因
# mean+2SD可以反映95%以上的观测值,设为mean+3SD,就可以反映97%以上的观测
logFC_cutoff <- with(DEG, mean(abs(logFC)) + 2*sd(abs(logFC)))
# 首先判断p值和logFC的绝对值是不是达到了设定的阈值,如果是则进行下一步判断,如果不是则返回NOT
# 然后判断logFC与阈值的大小关系,返回UP或DOWN
DEG$result <- as.factor(ifelse(DEG$P.Value < 0.05 & abs(DEG$logFC) > logFC_cutoff,
ifelse(DEG$logFC >logFC_cutoff, 'UP', 'DOWN'), 'NOT')
)
# 设置火山图标题
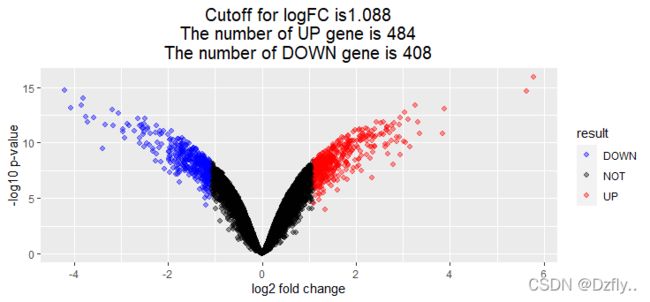
this_tile <- paste0('Cutoff for logFC is', round(logFC_cutoff, 3),
'\nThe number of UP gene is ', nrow(DEG[DEG$result == 'UP', ]),
'\nThe number of DOWN gene is ', nrow(DEG[DEG$result == 'DOWN', ]))
this_tile
head(DEG)
library(ggplot2)
# 对p值进行对数转换绘制的图就像火山喷发一样更美观
# 设置一系列的美化条件
ggplot(data=DEG, aes(x=logFC, y=-log10(P.Value), color=result)) +
geom_point(alpha=0.4, size=1.75) +
theme_set(theme_set(theme_bw(base_size=20)))+
xlab("log2 fold change") + ylab("-log10 p-value") +
ggtitle( this_tile ) + theme(plot.title = element_text(size=15,hjust = 0.5))+
scale_colour_manual(values = c('blue','black','red'))
# blue对应DOWN,black对应NOT,red对应UP
save(exprSet, group_list, nrDEG, DEG, file = 'GSE42872_DEG.Rdata')
?topTable :Value
DEG中的行变量对应的说明
A dataframe with a row for the number top genes and the following columns:
genelist:one or more columns of probe annotation, if genelist was included as input
logFC:estimate of the log2-fold-change corresponding to the effect or contrast (for topTableF there may be several columns of log-fold-changes)
CI.L:left limit of confidence interval for logFC (if confint=TRUE or confint is numeric)
CI.R:right limit of confidence interval for logFC (if confint=TRUE or confint is numeric)
AveExpr:average log2-expression for the probe over all arrays and channels, same as Amean in the MarrayLM object
t:moderated t-statistic (omitted for topTableF)
F:moderated F-statistic (omitted for topTable unless more than one coef is specified)
P.Value:raw p-value
adj.P.Value:adjusted p-value or q-value
B:log-odds that the gene is differentially expressed (omitted for topTreat)
STEP3:KEGG数据库注释
生信技能树:差异分析得到的结果注释一文就够
差异分析通过自定义的阈值挑选了有统计学显著的基因列表,我们需要对它们进行注释才能了解其功能,最常见的就是GO/KEGG数据库注释,当然也可以使用Reactome和Msigdb数据库来进行注释。最常见的注释方法就是超几何分布检验。
load('GSE42872_DEG.Rdata')
suppressPackageStartupMessages(library(clusterProfiler))
suppressPackageStartupMessages(library(org.Hs.eg.db))
# 这里可以 ?+函数名 看一下各个函数的帮助文档
# 注意函数输入数据的格式,按照要求修改数据的格式
gene <- head(rownames(nrDEG), 1000)
# bitr():Biological Id TRanslator
gene.df <- bitr(gene, fromType = "SYMBOL",
toType = c("ENSEMBL", "ENTREZID"),
OrgDb = org.Hs.eg.db)
head(gene.df)
# SYMBOL ENSEMBL ENTREZID
# 1 CD36 ENSG00000135218 948
# 2 DUSP6 ENSG00000139318 1848
# 3 DCT ENSG00000080166 1638
# 4 SPRY2 ENSG00000136158 10253
# 5 MOXD1 ENSG00000079931 26002
# 6 ETV4 ENSG00000175832 2118
# KEGG pathway analysis
# enrichKEGG():Given a vector of genes, this function will return the enrichment KEGG categories with FDR control.
kk <- enrichKEGG(gene = gene.df$ENTREZID, organism = "hsa",
pvalueCutoff = 0.05)
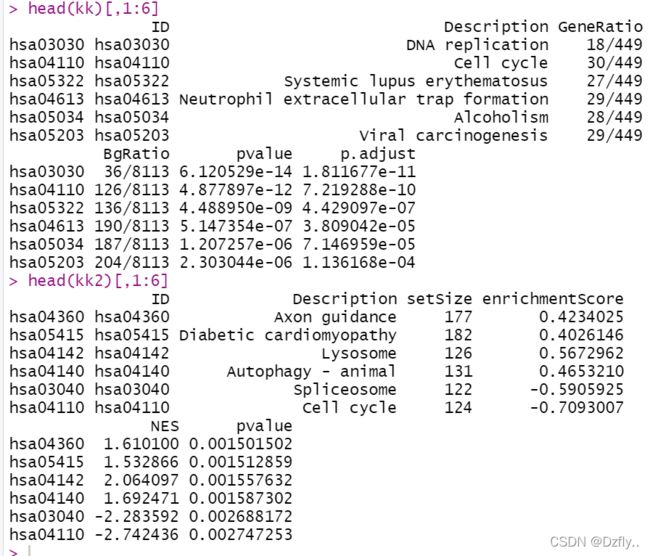
head(kk)[,1:6]
# kk2之前的所有操作,都是为了获得跟head(geneList)格式一样的数据
data(geneList, package = "DOSE")
boxplot(geneList)
head(geneList)
# 4312 8318 10874 55143 55388 991
# 4.572613 4.514594 4.418218 4.144075 3.876258 3.677857
boxplot(nrDEG$logFC)
geneList <- nrDEG$logFC
names(geneList) <- rownames(nrDEG)
head(geneList)
# CD36 DUSP6 DCT SPRY2 MOXD1 ETV4
# 5.780170 -4.212683 5.633027 -3.801663 3.263063 -3.843247
gene.symbol <- bitr(names(geneList), fromType = "SYMBOL",
toType = c("ENSEMBL", "ENTREZID"),
OrgDb = org.Hs.eg.db)
head(gene.symbol)
tmp <- data.frame(SYMBOL = names(geneList),
logFC = as.numeric(geneList))
tmp <- merge(tmp, gene.symbol, by = 'SYMBOL')
geneList <- tmp$logFC
names(geneList) <- tmp$ENTREZID
head(geneList)
# 29974 2 144568 127550 53947 51146
# -0.0490000 0.2959367 -0.1226300 -0.3733300 -0.4037100 -0.1646833
# gseKEGG要求genelist排好序
geneList <- sort(geneList, decreasing = T)
# gseKEGG():Gene Set Enrichment Analysis of KEGG
kk2 <- gseKEGG(geneList = geneList,
organism = 'hsa',
nPerm = 1000,
minGSSize = 120,
pvalueCutoff = 0.05,
verbose = FALSE)
head(kk2)[,1:6]
# visualize analyzing result of GSEA
# 图的结果看不懂...
gseaplot(kk2, geneSetID = "hsa04142")
完整代码
setwd(dir = "geo_learn/")
##############
### STEP 1 ###
##############
if(F){
suppressPackageStartupMessages(library(GEOquery))
gset <- getGEO('GSE42872', destdir=".",
AnnotGPL = F,
getGPL = F)
save(gset,file='GSE42872_gset.Rdata')
}
load('GSE42872_gset.Rdata')
exprSet <- exprs(gset[[1]])
pdata <- pData(gset[[1]])
group_list <- c(rep('control', 3), rep('case', 3))
# 以下操作等同于exprs(gset[[1]])
# a <- read.table("GSE42872_series_matrix.txt.gz",
# sep = "\t", quote = "", fill = T,
# comment.char = "!", header = T)
# rownames(a) <- a[,1]
# a <- a[,-1]
# BiocManager::install("hugene10sttranscriptcluster.db")
suppressPackageStartupMessages(library(hugene10sttranscriptcluster.db))
# 下载不到对应的R包时
# gpl <- getGE0('GPL6480', destdir=".")
# colnames(Table(gpl))##[1] 41108 17
# head(Table(gpL)[,c(1,6,7)])
# # you need to check this,which column do you need
# write.csv(Table(gpl)[,c(1,6,7)],"GPL6400.csv")
# ls("package:hugene10sttranscriptcluster.db")
ids <- toTable(hugene10sttranscriptclusterSYMBOL)
table(rownames(exprSet) %in% ids$probe_id)
# dim(exprSet)
exprSet <- exprSet[(rownames(exprSet) %in% ids$probe_id),]
# dim(exprSet)
ids <- ids[match(rownames(exprSet),ids$probe_id),]
# dim(ids)
tmp <- by(exprSet,ids$symbol,
function(x) rownames(x)[which.max(rowMeans(x))])
# tmp[1:20]
probes <- as.character(tmp)
exprSet <- exprSet[rownames(exprSet) %in% probes, ]
# dim(exprSet)
# dim(ids)
rownames(exprSet) <- ids[match(rownames(exprSet),ids$probe_id),2]
save(exprSet, group_list, file = 'GSE42872_new_exprSet.Rdata')
##############
### STEP 2 ###
##############
load('GSE42872_new_exprSet.Rdata')
# boxplot
library(reshape2)
m_exprSet <- melt(exprSet)
head(m_exprSet)
colnames(m_exprSet) <- c("symbol", "sample", "value")
head(m_exprSet)
m_exprSet$group <- rep(group_list, each = nrow(exprSet))
head(m_exprSet)
library(ggplot2)
ggplot(m_exprSet, aes(x = sample, y = value, fill = group)) + geom_boxplot()
# clustering
colnames(exprSet) <- paste(group_list,1:6,sep='')
hc <- hclust(dist(t(exprSet)))
nodePar <- list(lab.cex = 0.6, pch = c(NA, 19), cex = 0.7, col = "blue")
par(mar=c(5,5,5,10))
plot(as.dendrogram(hc), nodePar = nodePar, horiz = TRUE)
# limma
library(limma)
design <- model.matrix(~0 + factor(group_list))
colnames(design) <- levels(factor(group_list))
rownames(design) <- colnames(exprSet)
design
contrast.matrix <- makeContrasts("case-control" ,levels = design)
# contrast.matrix <- makeContrasts("control-case" ,levels = design)
contrast.matrix
##step1
fit <- lmFit(exprSet,design)
##step2
fit2 <- contrasts.fit(fit, contrast.matrix)
# Methods of assessing differential expression
fit2 <- eBayes(fit2)
##step3
# For topTable, fit should be an object of class MArrayLM as produced by lmFit and eBayes.
tempOutput <- topTable(fit2, coef=1, n=Inf)
nrDEG <- na.omit(tempOutput)
head(nrDEG)
## volcano plot
DEG <- nrDEG
logFC_cutoff <- with(DEG, mean(abs(logFC)) + 2*sd(abs(logFC)))
DEG$result <- as.factor(ifelse(DEG$P.Value < 0.05 & abs(DEG$logFC) > logFC_cutoff,
ifelse(DEG$logFC >logFC_cutoff, 'UP', 'DOWN'), 'NOT')
)
this_tile <- paste0('Cutoff for logFC is', round(logFC_cutoff, 3),
'\nThe number of UP gene is ', nrow(DEG[DEG$result == 'UP', ]),
'\nThe number of DOWN gene is ', nrow(DEG[DEG$result == 'DOWN', ]))
this_tile
head(DEG)
library(ggplot2)
ggplot(data=DEG, aes(x=logFC, y=-log10(P.Value), color=result)) +
geom_point(alpha=0.4, size=1.75) +
theme_set(theme_set(theme_bw(base_size=20)))+
xlab("log2 fold change") + ylab("-log10 p-value") +
ggtitle( this_tile ) + theme(plot.title = element_text(size=15,hjust = 0.5))+
scale_colour_manual(values = c('blue','black','red'))
save(exprSet, group_list, nrDEG, DEG, file = 'GSE42872_DEG.Rdata')
##############
### STEP 3 ###
##############
load('GSE42872_DEG.Rdata')
suppressPackageStartupMessages(library(clusterProfiler))
suppressPackageStartupMessages(library(org.Hs.eg.db))
gene <- head(rownames(nrDEG), 1000)
# bitr():Biological Id TRanslator
gene.df <- bitr(gene, fromType = "SYMBOL",
toType = c("ENSEMBL", "ENTREZID"),
OrgDb = org.Hs.eg.db)
head(gene.df)
# KEGG pathway analysis
kk <- enrichKEGG(gene = gene.df$ENTREZID, organism = "hsa",
pvalueCutoff = 0.05)
head(kk)[,1:6]
data(geneList, package = "DOSE")
boxplot(geneList)
head(geneList)
boxplot(nrDEG$logFC)
geneList <- nrDEG$logFC
names(geneList) <- rownames(nrDEG)
head(geneList)
gene.symbol <- bitr(names(geneList), fromType = "SYMBOL",
toType = c("ENSEMBL", "ENTREZID"),
OrgDb = org.Hs.eg.db)
head(gene.symbol)
tmp <- data.frame(SYMBOL = names(geneList),
logFC = as.numeric(geneList))
tmp <- merge(tmp, gene.symbol, by = 'SYMBOL')
geneList <- tmp$logFC
names(geneList) <- tmp$ENTREZID
head(geneList)
geneList <- sort(geneList, decreasing = T)
kk2 <- gseKEGG(geneList = geneList,
organism = 'hsa',
nPerm = 1000,
minGSSize = 120,
pvalueCutoff = 0.05,
verbose = FALSE)
head(kk2)[,1:6]
gseaplot(kk2, geneSetID = "hsa04142")