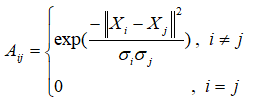相似度矩阵的几种构造方式(附代码)
标题
在谱聚类中,常将数据看做空间上的点,点之间用边连接起来,构造无向权重图 G = ( V , E ) \mathbf{G}=(\mathbf{V},\mathbf{E}) G=(V,E),其中 V = { v 1 , v 2 , . . . , v n } \mathbf{V}=\{v_1, v_2, ..., v_n\} V={v1,v2,...,vn}为定点集合,E为边集合。对任意一个顶点Vi,度di表示与它相连的所有边权重之和,W(i,j)表示Vi和Vj之间的边权重,那么![]() 那么对于n个顶点,可构造 n ∗ n n*n n∗n的度矩阵D=diag(d1,d2,…,dn)。同时,利用所有顶点之间的权重,得邻接矩阵(相似度矩阵)W。
那么对于n个顶点,可构造 n ∗ n n*n n∗n的度矩阵D=diag(d1,d2,…,dn)。同时,利用所有顶点之间的权重,得邻接矩阵(相似度矩阵)W。
常见的邻接矩阵W的构造方法有3种,分别是 ε \varepsilon ε-邻近、k-邻近、全连接法。
本文主要基于高斯核的全连接法构造邻接矩阵W。 ε \varepsilon ε-邻近、k-邻近的具体构造方法见这篇文章。
目录:
-
-
- 当 σ \sigma σ是单值(single scale)
-
-
- ① 纯手工计算
- ② 蔡登的constructW.m
-
- 当 σ \sigma σ是局部值(local scaling parameter)
-
-
- ① “self-tuning spectral clustering”论文代码
- ②自编代码selftuning2.m
-
-
已知 X = [ x 1 , x 2 , ⋯ , x n ] ∈ R n × d \mathbf{X}=[\mathbf{x}_1,\mathbf{x}_2,\cdots,\mathbf{x}_n]\in\mathbb{R}^{n\times d} X=[x1,x2,⋯,xn]∈Rn×d,使用高斯核RBF作为核函数,有:
 其中, σ \sigma σ表示数据点的邻域宽度。
其中, σ \sigma σ表示数据点的邻域宽度。
目前,对于 σ \sigma σ有两种不同输入,一种由人工选取,作为固定值(single scale parameter,单值参数)输入,另一种由self-tuning算法提出的自动选取方法,即 σ \sigma σ是一个变量(local scaling parameter,局部值),对于不同的样本点, σ \sigma σ不同。
当 σ \sigma σ是单值(single scale)
① 纯手工计算
% 输入X,sigma
[row,col] = size(X);
W = zeros(row,row);
index_all = zeros(row,row);
for i = 1:row % 全连接构造相似度矩阵W
for j=1:row
if i ~= j
W(i,j) = exp((-sum((X(i,:)-X(j,:)).^2))/(2*sigma.^2));
end
end
end
Affinity = W;
② 蔡登的constructW.m
完整代码见:github
注意,要使用全连接图,option.k = 0.
运行结果 W 与‘’① 纯手工计算‘’结果应一致.(W与后文的邻接矩阵/亲和矩阵A 相同)
function W = constructW(fea,options)
% Usage:
% W = constructW(fea,options)
%
% fea: Rows of vectors of data points. Each row is x_i
% options: Struct value in Matlab. The fields in options that can be set:
%
% NeighborMode - Indicates how to construct the graph. Choices
% are: [Default 'KNN']
% 'KNN' - k = 0
% Complete graph
% k > 0
% Put an edge between two nodes if and
% only if they are among the k nearst
% neighbors of each other. You are
% required to provide the parameter k in
% the options. Default k=5.
% 'Supervised' - k = 0
% Put an edge between two nodes if and
% only if they belong to same class.
% k > 0
% Put an edge between two nodes if
% they belong to same class and they
% are among the k nearst neighbors of
% each other.
% Default: k=0
% You are required to provide the label
% information gnd in the options.
%
% WeightMode - Indicates how to assign weights for each edge
% in the graph. Choices are:
% 'Binary' - 0-1 weighting. Every edge receiveds weight
% of 1.
% 'HeatKernel' - If nodes i and j are connected, put weight
% W_ij = exp(-norm(x_i - x_j)/2t^2). You are
% required to provide the parameter t. [Default One]
% 'Cosine' - If nodes i and j are connected, put weight
% cosine(x_i,x_j).
%
% k - The parameter needed under 'KNN' NeighborMode.
% Default will be 5.
% gnd - The parameter needed under 'Supervised'
% NeighborMode. Colunm vector of the label
% information for each data point.
% bLDA - 0 or 1. Only effective under 'Supervised'
% NeighborMode. If 1, the graph will be constructed
% to make LPP exactly same as LDA. Default will be
% 0.
% t - The parameter needed under 'HeatKernel'
% WeightMode. Default will be 1
% bNormalized - 0 or 1. Only effective under 'Cosine' WeightMode.
% Indicates whether the fea are already be
% normalized to 1. Default will be 0
% bSelfConnected - 0 or 1. Indicates whether W(i,i) == 1. Default 0
% if 'Supervised' NeighborMode & bLDA == 1,
% bSelfConnected will always be 1. Default 0.
% bTrueKNN - 0 or 1. If 1, will construct a truly kNN graph
% (Not symmetric!). Default will be 0. Only valid
% for 'KNN' NeighborMode
% For more details about the different ways to construct the W, please
% refer:
% Deng Cai, Xiaofei He and Jiawei Han, "Document Clustering Using
% Locality Preserving Indexing" IEEE TKDE, Dec. 2005.
%
%
% Written by Deng Cai (dengcai2 AT cs.uiuc.edu), April/2004, Feb/2006,
% May/2007
options = [];
options.NeighborMode = 'KNN';
options.k = 0; % 全连接图
options.WeightMode = 'HeatKernel';
options.t = 1; % Kernel sigma,要与‘① 纯手工计算’中设定的一致
W = constructW(X,options);
Affinity = full(W);
当 σ \sigma σ是局部值(local scaling parameter)
注意,指数部分的分母没有1/2
其中, X i K \mathbf{X_{iK}} XiK是 X i \mathbf{X_{i}} Xi样本点的第K个邻近点(在距离上)。
![]()
该思想源自作者Lihi Zelnik-Manor的论文《Self-Tuning Spectral Clustering》中的Section 2,论文的代码包在我的github上:ZPclustering.rar。
① “self-tuning spectral clustering”论文代码
核心代码文件:dist2aff.cpp / scale_dist.cpp / segment_image.m / test1.m
dist2aff.cpp :输入 distmatrix,X的欧式平方距离矩阵
sigma , 高斯核函数的参数
输出 : A ,sigle scale下的邻接矩阵,公式exp(-distmatrix./(sigma*sigma))
scale_dist.cpp :输入 distmatrix,X的欧式平方距离矩阵
k, sigma(i)取决于样本及其第k个邻近点的欧氏距离sigma(i)=||Xi-XiK||
输出 : A ,local scale下的邻接矩阵,公式exp(-distmatrix(i,j)./(sigma(i)*sigma(j)))
scale_dist.cpp代码内容补充解释:
LS(i,j) = sigma(i)*sigma(j)
D(i,j) = ||Xi-Xj||^2
D_LS(i,j) = ( ||Xi-Xj||^2 )/ (sigma(i)*sigma(j)) = LS(i,j)/D(i,j)
A_LS(i,j) = exp{( ||Xi-Xj||^2 )/ (sigma(i)*sigma(j))} = exp(D_LS(i,j))
(以下若遇到解决错误使用mex未找到支持的编译器或SDK问题,根据Matlab提示网站查看details,下载sdk 7.1 和.Net 4.0等操作,部分安装指导:参考博客1的3.1部分、参考博客2,若编译成功显示如:)
我整理的相似度矩阵构造代码:main_Run.m,如下:
mex dist2aff.cpp ;
mex scale_dist.cpp ;
%%%%%%%%%%%%%%%%% Build affinity matrices
% the equation of kernel function is exp(-dist(i,j)/sigma1*simga2),for
% single-scale affinity construction,sigma1 equal to sigma2 ,while for
% automatic-selftuning affinity construction ,the value of sigma1,2 is
% dependent of the the k'th neughbor of Xi.
% input:X ,scale
% output:A , A_LS
%
%***** Ayla Huang ****
scale = 0.1; %% parameter sigma in kernel
D = dist2(X,X); %% Euclidean square distance
A = exp(-D/(scale^2)); %% Standard affinity matrix (single scale)
k = 5; %% the number of neighbor, the k'th neighbor for selftuning
[D_LS,A_LS,LS] = scale_dist(D,k); %% Locally scaled affinity matrix (automatic selftuning)
clear D_LS; clear LS;
% Zero out diagonal
% the diag-element in A and A_LS is 1 ,now turn it to zero.
ZERO_DIAG = ~eye(size(X,1));
A = A.*ZERO_DIAG;
A_LS = A_LS.*ZERO_DIAG;
②自编代码selftuning2.m
原理相同,用纯Matlab实现。
注意A(i,j) = exp((-sum((X(i,:)-X(j,:)).^2))/(sigma(i)*sigma(j)));
运行结果 A 与‘’① “self-tuning spectral clustering”论文代码‘’结果一致.
function [A,An] = selftuning2(X,k)
%% load Data
% X = getData(data_name);
%% construct affinity matrix A
[n,~]=size(X);
% 全连接欧式距离矩阵dis_all
dis_all = L2_distance_1(X',X'); % 平方距离
dis_all(find(dis_all<0)) = 0;
dis_all = sqrt(dis_all);
% 使用KNN计算欧式距离最近的 '第k个'点sigma,保留n个样本的第k近距离结果sigma(n,1)
[dumb,idx] = sort(dis_all, 2); % sort each row
index=idx(:,k+1);
sigma = zeros(n,1);
for i=1:n
sigma(i)=sqrt(sum((X(i,:)-X(index(i),:)).^2));
end
%构造selftuning下的affinity矩阵A
A=zeros(n,n);
for i = 1:n
for j=1:n
if i ~= j
A(i,j) = exp((-sum((X(i,:)-X(j,:)).^2))/(sigma(i)*sigma(j)));
end
end
end
D = diag(sum(A,2));
Dn1 = D^(-0.5);
An = Dn1*A*Dn1; %前c个最大,An是拉普拉斯矩阵
clear Dn;
An = max(An,An');
function d = L2_distance_1(a,b)
% compute squared Euclidean distance
% ||A-B||^2 = ||A||^2 + ||B||^2 - 2*A'*B
% a,b: two matrices. each column is a data
% d: distance matrix of a and b
if (size(a,1) == 1)
a = [a; zeros(1,size(a,2))];
b = [b; zeros(1,size(b,2))];
end
aa=sum(a.*a); bb=sum(b.*b); ab=a'*b;
d = repmat(aa',[1 size(bb,2)]) + repmat(bb,[size(aa,2) 1]) - 2*ab;
d = real(d);
d = max(d,0);