Deepsort 算法的介绍
Deep-Sort 多目标跟踪算法原理和代码解析
deepsort是基于目标检测的多目标跟踪算法(Mutil-object Tracking),目标检测算法的优劣影响该算法跟踪的效果。
1.MOT算法的主要步骤
- 给定视频的初始帧
- 运行目标检测算法,例如YOLO、Faster R-CNN 、SSD等算法对视频每帧进行检测,获得检测边界框
- 根据检测边界框对图片进行裁剪获得检测目标,再依次对目标进行特征提取(表观特征或运动特征)
- 根据提取的特征,计算前后两帧的相似度矩阵(cost_metrix)
- 数据关联,为每个对象分配目标ID
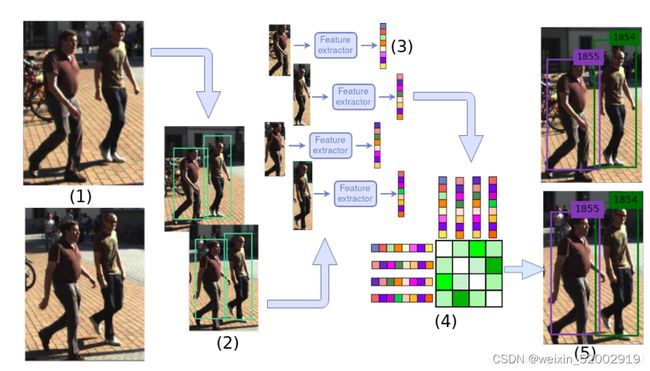
2.简述Sort算法流程
SORT算法是Deepsort算法的前身。其两个核心算法为卡尔曼滤波算法和匈牙利算法。
卡尔曼滤波算法:分为预测和更新两个部分,预测:根据t-1帧的运动状态预测t帧的运动状态,即位置和速度;更新:将t帧检测目标和t帧预测结果线性加权
匈牙利算法:解决线性分配问题。sklearn库有对应的linear_assignment方法。
SORT算法的流程为:
- 通过运行目标检测方法获得每帧目标检测框detections;
- 使用kalman filter 对前一帧的Tracks中每个track进行预测,得到当前帧track 的mean 和covariance,其中mean为目标的运动特征向量:[x,y,a,h,Vx,Vy,Va,Vh]
- 将detections和tracks进行IOU匹配,得到Matched tracks、Unmatched detections和Unmatched tracks
- kalman filter将Matched tracks状态更新;
- Unmatched detections 初始化为新的轨迹
- Unmatched tracks 删除
以下是Sort 算法流程图

3 Deep Sort
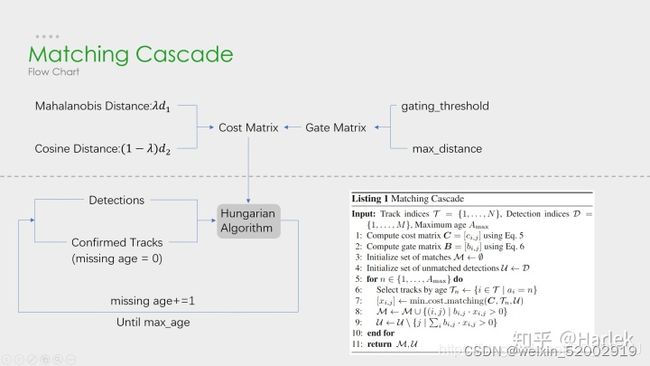
Deepsort 在sort的基础上添加了rein网络模型提取检测框的表观特征,并采用级联匹配(Matching Cascade),减少了目标ID switch次数。整体流程如下图:

- 运行kalman filter 对tracks预测;
*使用匈牙利算法对 detections与confirmed状态的tracks进行级联匹配,获得Unmatched tracks 、Unmatched detections和matched tracks; - 由tentative状态和Unmatched tracks &time_since_updata=1的track构成候选轨迹,与Unmatched detections使用匈牙利算法进行IOU匹配;
- kalman filter进行更新。
其上图的级联匹配展开为下图:(其中第二张图是我查看代码觉得deepsort级联匹配应该是该张图)
# tracker._match
def _match(self, detections):
def gated_metric(tracks, dets, track_indices, detection_indices):
features = np.array([dets[i].feature for i in detection_indices])
targets = np.array([tracks[i].track_id for i in track_indices])
cost_matrix = self.metric.distance(features, targets) #调用nn_matching.NearestNeighborDistanceMetric.distance函数求features与targets的余弦距离矩阵
cost_matrix = linear_assignment.gate_cost_matrix(
self.kf, cost_matrix, tracks, dets, track_indices,
detection_indices)
return cost_matrix
confirmed_tracks = [
i for i, t in enumerate(self.tracks) if t.is_confirmed()]
unconfirmed_tracks = [
i for i, t in enumerate(self.tracks) if not t.is_confirmed()]
# Associate confirmed tracks using appearance features.
matches_a, unmatched_tracks_a, unmatched_detections = \
linear_assignment.matching_cascade(
gated_metric, self.metric.matching_threshold, self.max_age,
self.tracks, detections, confirmed_tracks)
# linear_assignment.gate_cost_matrix
def gate_cost_matrix(
kf, cost_matrix, tracks, detections, track_indices, detection_indices,
gated_cost=INFTY_COST, only_position=False):
gating_dim = 2 if only_position else 4
gating_threshold = kalman_filter.chi2inv95[gating_dim]
measurements = np.asarray(
[detections[i].to_xyah() for i in detection_indices])
for row, track_idx in enumerate(track_indices):
track = tracks[track_idx]
gating_distance = kf.gating_distance(
track.mean, track.covariance, measurements, only_position)
cost_matrix[row, gating_distance > gating_threshold] = gated_cost
return cost_matrix
def matching_cascade(
distance_metric, max_distance, cascade_depth, tracks, detections,
track_indices=None, detection_indices=None):
if track_indices is None:
track_indices = list(range(len(tracks)))
if detection_indices is None:
detection_indices = list(range(len(detections)))
unmatched_detections = detection_indices
matches = []
for level in range(cascade_depth):
if len(unmatched_detections) == 0: # No detections left
break
track_indices_l = [
k for k in track_indices
if tracks[k].time_since_update == 1 + level
]
if len(track_indices_l) == 0: # Nothing to match at this level
continue
matches_l, _, unmatched_detections = \
min_cost_matching(
distance_metric, max_distance, tracks, detections,
track_indices_l, unmatched_detections)
matches += matches_l
unmatched_tracks = list(set(track_indices) - set(k for k, _ in matches))
return matches, unmatched_tracks, unmatched_detections
级联匹配具体流程:
- 首先代价矩阵是由表观特征计算的余弦距离矩阵,运动特征计算马氏距离矩阵;
- 马氏距离矩阵与gating_threshold、max_distance 比较,使用gated_metrix对代价矩阵进行修正
- 没有匹配miss的tracks优先与detections进行匈牙利算法数据关联,循环max_age=70次
通过这部分处理,可以重新将被遮挡目标找回,降低被遮挡然后再出现的目标发生的ID Switch次数。
4.deepsort代码解析
论文中提供的代码是如下地址: https://github.com/mikel-brostrom/Yolov5_DeepSort_Pytorch
仅对以下的deepsort核心代码进行解析,本次不涉及目标检测部分。

4.1 Kalman Filter 模块

kalman filter的详情介绍可以参考以下链接:
[https://blog.csdn.net/u010720661/article/details/63253509]
# vim: expandtab:ts=4:sw=4
import numpy as np
import scipy.linalg
"""
Table for the 0.95 quantile of the chi-square distribution with N degrees of
freedom (contains values for N=1, ..., 9). Taken from MATLAB/Octave's chi2inv
function and used as Mahalanobis gating threshold.
"""
chi2inv95 = {
1: 3.8415,
2: 5.9915,
3: 7.8147,
4: 9.4877,
5: 11.070,
6: 12.592,
7: 14.067,
8: 15.507,
9: 16.919}
class KalmanFilter(object):
"""
A simple Kalman filter for tracking bounding boxes in image space.
The 8-dimensional state space
x, y, a, h, vx, vy, va, vh
contains the bounding box center position (x, y), aspect ratio a, height h,
and their respective velocities.
Object motion follows a constant velocity model. The bounding box location
(x, y, a, h) is taken as direct observation of the state space (linear
observation model).
"""
def __init__(self):
ndim, dt = 4, 1.
# Create Kalman filter model matrices(状态转移矩阵).
self._motion_mat = np.eye(2 * ndim, 2 * ndim)
for i in range(ndim):
self._motion_mat[i, ndim + i] = dt
self._update_mat = np.eye(ndim, 2 * ndim) # 测量矩阵
# Motion and observation uncertainty are chosen relative to the current
# state estimate. These weights control the amount of uncertainty in
# the model. This is a bit hacky.
self._std_weight_position = 1. / 20
self._std_weight_velocity = 1. / 160
def initiate(self, measurement):
"""Create track from unassociated measurement.
Parameters
----------
measurement : ndarray
Bounding box coordinates (x, y, a, h) with center position (x, y),
aspect ratio a, and height h.
Returns
-------
(ndarray, ndarray)
Returns the mean vector (8 dimensional) and covariance matrix (8x8
dimensional) of the new track. Unobserved velocities are initialized
to 0 mean.
"""
mean_pos = measurement
mean_vel = np.zeros_like(mean_pos)
mean = np.r_[mean_pos, mean_vel]
std = [
2 * self._std_weight_position * measurement[3],
2 * self._std_weight_position * measurement[3],
1e-2,
2 * self._std_weight_position * measurement[3],
10 * self._std_weight_velocity * measurement[3],
10 * self._std_weight_velocity * measurement[3],
1e-5,
10 * self._std_weight_velocity * measurement[3]]
covariance = np.diag(np.square(std))
return mean, covariance
def predict(self, mean, covariance):
"""Run Kalman filter prediction step.
Parameters
----------
mean : ndarray
The 8 dimensional mean vector of the object state at the previous
time step.
covariance : ndarray
The 8x8 dimensional covariance matrix of the object state at the
previous time step.
Returns
-------
(ndarray, ndarray)
Returns the mean vector and covariance matrix of the predicted
state. Unobserved velocities are initialized to 0 mean.
"""
std_pos = [
self._std_weight_position * mean[3],
self._std_weight_position * mean[3],
1e-2,
self._std_weight_position * mean[3]]
std_vel = [
self._std_weight_velocity * mean[3],
self._std_weight_velocity * mean[3],
1e-5,
self._std_weight_velocity * mean[3]]
motion_cov = np.diag(np.square(np.r_[std_pos, std_vel]))
mean = np.dot(self._motion_mat, mean)
covariance = np.linalg.multi_dot((
self._motion_mat, covariance, self._motion_mat.T)) + motion_cov
return mean, covariance
def project(self, mean, covariance):
"""Project state distribution to measurement space.
Parameters
----------
mean : ndarray
The state's mean vector (8 dimensional array).
covariance : ndarray
The state's covariance matrix (8x8 dimensional).
Returns
-------
(ndarray, ndarray)
Returns the projected mean and covariance matrix of the given state
estimate.
"""
std = [
self._std_weight_position * mean[3],
self._std_weight_position * mean[3],
1e-1,
self._std_weight_position * mean[3]]
innovation_cov = np.diag(np.square(std)) # 噪声矩阵
mean = np.dot(self._update_mat, mean)
covariance = np.linalg.multi_dot((
self._update_mat, covariance, self._update_mat.T))
return mean, covariance + innovation_cov
def update(self, mean, covariance, measurement):
"""Run Kalman filter correction step.
Parameters
----------
mean : ndarray
The predicted state's mean vector (8 dimensional).
covariance : ndarray
The state's covariance matrix (8x8 dimensional).
measurement : ndarray
The 4 dimensional measurement vector (x, y, a, h), where (x, y)
is the center position, a the aspect ratio, and h the height of the
bounding box.
Returns
-------
(ndarray, ndarray)
Returns the measurement-corrected state distribution.
"""
projected_mean, projected_cov = self.project(mean, covariance)
chol_factor, lower = scipy.linalg.cho_factor(
projected_cov, lower=True, check_finite=False)
kalman_gain = scipy.linalg.cho_solve(
(chol_factor, lower), np.dot(covariance, self._update_mat.T).T,
check_finite=False).T
innovation = measurement - projected_mean
new_mean = mean + np.dot(innovation, kalman_gain.T)
new_covariance = covariance - np.linalg.multi_dot((
kalman_gain, projected_cov, kalman_gain.T))
return new_mean, new_covariance
def gating_distance(self, mean, covariance, measurements,
only_position=False):
"""Compute gating distance between state distribution and measurements.
A suitable distance threshold can be obtained from `chi2inv95`. If
`only_position` is False, the chi-square distribution has 4 degrees of
freedom, otherwise 2.
Parameters
----------
mean : ndarray
Mean vector over the state distribution (8 dimensional).
covariance : ndarray
Covariance of the state distribution (8x8 dimensional).
measurements : ndarray
An Nx4 dimensional matrix of N measurements, each in
format (x, y, a, h) where (x, y) is the bounding box center
position, a the aspect ratio, and h the height.
only_position : Optional[bool]
If True, distance computation is done with respect to the bounding
box center position only.
Returns
-------
ndarray
Returns an array of length N, where the i-th element contains the
squared Mahalanobis distance between (mean, covariance) and
`measurements[i]`.
"""
mean, covariance = self.project(mean, covariance)
if only_position:
mean, covariance = mean[:2], covariance[:2, :2]
measurements = measurements[:, :2]
cholesky_factor = np.linalg.cholesky(covariance)
d = measurements - mean
z = scipy.linalg.solve_triangular(
cholesky_factor, d.T, lower=True, check_finite=False,
overwrite_b=True)
squared_maha = np.sum(z * z, axis=0)
return squared_maha
4.2 detection 模块
import numpy as np
class Detection(object):
"""
This class represents a bounding box detection in a single image.
Parameters
----------
tlwh : array_like
Bounding box in format `(x, y, w, h)`.
confidence : float
Detector confidence score.
feature : array_like
A feature vector that describes the object contained in this image.
Attributes
----------
tlwh : ndarray
Bounding box in format `(top left x, top left y, width, height)`.
confidence : ndarray
Detector confidence score.
feature : ndarray | NoneType
A feature vector that describes the object contained in this image.
"""
def __init__(self, tlwh, confidence, feature):
self.tlwh = np.asarray(tlwh, dtype=np.float)
self.confidence = float(confidence)
self.feature = np.asarray(feature, dtype=np.float32)
def to_tlbr(self):
"""Convert bounding box to format `(min x, min y, max x, max y)`, i.e.,
`(top left, bottom right)`.
"""
ret = self.tlwh.copy()
ret[2:] += ret[:2]
return ret
def to_xyah(self):
"""Convert bounding box to format `(center x, center y, aspect ratio,
height)`, where the aspect ratio is `width / height`.
"""
ret = self.tlwh.copy()
ret[:2] += ret[2:] / 2
ret[2] /= ret[3]
return ret
4.3 nn_matching 模块
import numpy as np
def _pdist(a, b):
"""Compute pair-wise squared distance between points in `a` and `b`.
Parameters
----------
a : array_like
An NxM matrix of N samples of dimensionality M.
b : array_like
An LxM matrix of L samples of dimensionality M.
Returns
-------
ndarray
Returns a matrix of size len(a), len(b) such that eleement (i, j)
contains the squared distance between `a[i]` and `b[j]`.
"""
a, b = np.asarray(a), np.asarray(b)
if len(a) == 0 or len(b) == 0:
return np.zeros((len(a), len(b)))
a2, b2 = np.square(a).sum(axis=1), np.square(b).sum(axis=1)
r2 = -2. * np.dot(a, b.T) + a2[:, None] + b2[None, :]
r2 = np.clip(r2, 0., float(np.inf))
return r2
def _cosine_distance(a, b, data_is_normalized=False):
"""Compute pair-wise cosine distance between points in `a` and `b`.
Parameters
----------
a : array_like
An NxM matrix of N samples of dimensionality M.
b : array_like
An LxM matrix of L samples of dimensionality M.
data_is_normalized : Optional[bool]
If True, assumes rows in a and b are unit length vectors.
Otherwise, a and b are explicitly normalized to lenght 1.
Returns
-------
ndarray
Returns a matrix of size len(a), len(b) such that eleement (i, j)
contains the cosine distance between `a[i]` and `b[j]`.
"""
if not data_is_normalized:
a = np.asarray(a) / np.linalg.norm(a, axis=1, keepdims=True)
b = np.asarray(b) / np.linalg.norm(b, axis=1, keepdims=True)
return 1. - np.dot(a, b.T)
def _nn_euclidean_distance(x, y):
""" Helper function for nearest neighbor distance metric (Euclidean).
Parameters
----------
x : ndarray
A matrix of N row-vectors (sample points).
y : ndarray
A matrix of M row-vectors (query points).
Returns
-------
ndarray
A vector of length M that contains for each entry in `y` the
smallest Euclidean distance to a sample in `x`.
"""
distances = _pdist(x, y)
return np.maximum(0.0, distances.min(axis=0))
def _nn_cosine_distance(x, y):
""" Helper function for nearest neighbor distance metric (cosine).
Parameters
----------
x : ndarray
A matrix of N row-vectors (sample points).
y : ndarray
A matrix of M row-vectors (query points).
Returns
-------
ndarray
A vector of length M that contains for each entry in `y` the
smallest cosine distance to a sample in `x`.
"""
distances = _cosine_distance(x, y)
return distances.min(axis=0)
class NearestNeighborDistanceMetric(object):
"""
A nearest neighbor distance metric that, for each target, returns
the closest distance to any sample that has been observed so far.
Parameters
----------
metric : str
Either "euclidean" or "cosine".
matching_threshold: float
The matching threshold. Samples with larger distance are considered an
invalid match.
budget : Optional[int]
If not None, fix samples per class to at most this number. Removes
the oldest samples when the budget is reached.
Attributes
----------
samples : Dict[int -> List[ndarray]]
A dictionary that maps from target identities to the list of samples
that have been observed so far.
"""
def __init__(self, metric, matching_threshold, budget=None):
if metric == "euclidean":
self._metric = _nn_euclidean_distance
elif metric == "cosine":
self._metric = _nn_cosine_distance
else:
raise ValueError(
"Invalid metric; must be either 'euclidean' or 'cosine'")
self.matching_threshold = matching_threshold
self.budget = budget
self.samples = {}
def partial_fit(self, features, targets, active_targets):
"""Update the distance metric with new data.
Parameters
----------
features : ndarray
An NxM matrix of N features of dimensionality M.
targets : ndarray
An integer array of associated target identities.
active_targets : List[int]
A list of targets that are currently present in the scene.
"""
for feature, target in zip(features, targets):
self.samples.setdefault(target, []).append(feature)
if self.budget is not None:
self.samples[target] = self.samples[target][-self.budget:]
self.samples = {k: self.samples[k] for k in active_targets}
def distance(self, features, targets):
"""Compute distance between features and targets.
Parameters
----------
features : ndarray
An NxM matrix of N features of dimensionality M.
targets : List[int]
A list of targets to match the given `features` against.
Returns
-------
ndarray
Returns a cost matrix of shape len(targets), len(features), where
element (i, j) contains the closest squared distance between
`targets[i]` and `features[j]`.
"""
cost_matrix = np.zeros((len(targets), len(features)))
for i, target in enumerate(targets):
cost_matrix[i, :] = self._metric(self.samples[target], features)
return cost_matrix
4.4 IOU_matching 模块
# vim: expandtab:ts=4:sw=4
from __future__ import absolute_import
import numpy as np
from . import linear_assignment
def iou(bbox, candidates):
"""Computer intersection over union.
Parameters
----------
bbox : ndarray
A bounding box in format `(top left x, top left y, width, height)`.
candidates : ndarray
A matrix of candidate bounding boxes (one per row) in the same format
as `bbox`.
Returns
-------
ndarray
The intersection over union in [0, 1] between the `bbox` and each
candidate. A higher score means a larger fraction of the `bbox` is
occluded by the candidate.
"""
bbox_tl, bbox_br = bbox[:2], bbox[:2] + bbox[2:]
candidates_tl = candidates[:, :2]
candidates_br = candidates[:, :2] + candidates[:, 2:]
tl = np.c_[np.maximum(bbox_tl[0], candidates_tl[:, 0])[:, np.newaxis],
np.maximum(bbox_tl[1], candidates_tl[:, 1])[:, np.newaxis]]
br = np.c_[np.minimum(bbox_br[0], candidates_br[:, 0])[:, np.newaxis],
np.minimum(bbox_br[1], candidates_br[:, 1])[:, np.newaxis]]
wh = np.maximum(0., br - tl)
area_intersection = wh.prod(axis=1)
area_bbox = bbox[2:].prod()
area_candidates = candidates[:, 2:].prod(axis=1)
return area_intersection / (area_bbox + area_candidates - area_intersection)
def iou_cost(tracks, detections, track_indices=None,
detection_indices=None):
"""An intersection over union distance metric.
Parameters
----------
tracks : List[deep_sort.track.Track]
A list of tracks.
detections : List[deep_sort.detection.Detection]
A list of detections.
track_indices : Optional[List[int]]
A list of indices to tracks that should be matched. Defaults to
all `tracks`.
detection_indices : Optional[List[int]]
A list of indices to detections that should be matched. Defaults
to all `detections`.
Returns
-------
ndarray
Returns a cost matrix of shape
len(track_indices), len(detection_indices) where entry (i, j) is
`1 - iou(tracks[track_indices[i]], detections[detection_indices[j]])`.
"""
if track_indices is None:
track_indices = np.arange(len(tracks))
if detection_indices is None:
detection_indices = np.arange(len(detections))
cost_matrix = np.zeros((len(track_indices), len(detection_indices)))
for row, track_idx in enumerate(track_indices):
if tracks[track_idx].time_since_update > 1:
cost_matrix[row, :] = linear_assignment.INFTY_COST
continue
bbox = tracks[track_idx].to_tlwh()
candidates = np.asarray(
[detections[i].tlwh for i in detection_indices])
cost_matrix[row, :] = 1. - iou(bbox, candidates)
return cost_matrix
4.5 track 模块
# vim: expandtab:ts=4:sw=4
class TrackState:
"""
Enumeration type for the single target track state. Newly created tracks are
classified as `tentative` until enough evidence has been collected. Then,
the track state is changed to `confirmed`. Tracks that are no longer alive
are classified as `deleted` to mark them for removal from the set of active
tracks.
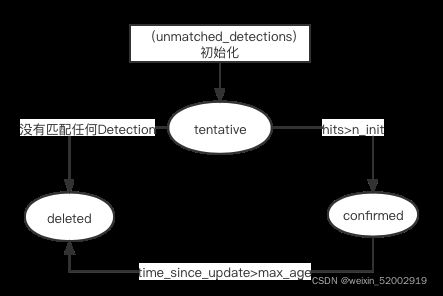
单个轨迹的状态:confirmed、tentative、deleted
"""
Tentative = 1
Confirmed = 2
Deleted = 3
class Track:
"""
A single target track with state space `(x, y, a, h)` and associated
velocities, where `(x, y)` is the center of the bounding box, `a` is the
aspect ratio and `h` is the height.
一个轨迹的信息,包含(x,y,a,h) 和速度,其中 (x,y)是框的中心点,a 是宽高比,h:框高
Parameters
----------
mean : ndarray
Mean vector of the initial state distribution.
covariance : ndarray
Covariance matrix of the initial state distribution.
track_id : int
A unique track identifier.
n_init : int
Number of consecutive detections before the track is confirmed. The
track state is set to `Deleted` if a miss occurs within the first
`n_init` frames.
max_age : int
The maximum number of consecutive misses before the track state is
set to `Deleted`.
feature : Optional[ndarray]
Feature vector of the detection this track originates from. If not None,
this feature is added to the `features` cache.
Attributes
----------
mean : ndarray
Mean vector of the initial state distribution.
covariance : ndarray
Covariance matrix of the initial state distribution.
track_id : int
A unique track identifier.
hits : int
Total number of measurement updates.
age : int
Total number of frames since first occurance.
time_since_update : int
Total number of frames since last measurement update.
state : TrackState
The current track state.
features : List[ndarray]
A cache of features. On each measurement update, the associated feature
vector is added to this list.
"""
def __init__(self, mean, covariance, track_id, n_init, max_age,
feature=None):
self.mean = mean
self.covariance = covariance
self.track_id = track_id
self.hits = 1
self.age = 1
self.time_since_update = 0
self.state = TrackState.Tentative
self.features = []
if feature is not None:
self.features.append(feature)
self._n_init = n_init
self._max_age = max_age
def to_tlwh(self):
"""Get current position in bounding box format `(top left x, top left y,
width, height)`.
Returns
-------
ndarray
The bounding box.
"""
ret = self.mean[:4].copy()
ret[2] *= ret[3]
ret[:2] -= ret[2:] / 2
return ret
def to_tlbr(self):
"""Get current position in bounding box format `(min x, miny, max x,
max y)`.
Returns
-------
ndarray
The bounding box.
"""
ret = self.to_tlwh()
ret[2:] = ret[:2] + ret[2:]
return ret
def increment_age(self):
self.age += 1
self.time_since_update += 1
def predict(self, kf):
"""Propagate the state distribution to the current time step using a
Kalman filter prediction step.
Parameters
----------
kf : kalman_filter.KalmanFilter
The Kalman filter.
"""
self.mean, self.covariance = kf.predict(self.mean, self.covariance)
self.increment_age()
def update(self, kf, detection):
"""Perform Kalman filter measurement update step and update the feature
cache.
Parameters
----------
kf : kalman_filter.KalmanFilter
The Kalman filter.
detection : Detection
The associated detection.
"""
self.mean, self.covariance = kf.update(
self.mean, self.covariance, detection.to_xyah())
self.features.append(detection.feature)
self.hits += 1
self.time_since_update = 0
if self.state == TrackState.Tentative and self.hits >= self._n_init:
self.state = TrackState.Confirmed
def mark_missed(self):
"""Mark this track as missed (no association at the current time step).
"""
if self.state == TrackState.Tentative:
self.state = TrackState.Deleted
elif self.time_since_update > self._max_age:
self.state = TrackState.Deleted
def is_tentative(self):
"""Returns True if this track is tentative (unconfirmed).
"""
return self.state == TrackState.Tentative
def is_confirmed(self):
"""Returns True if this track is confirmed."""
return self.state == TrackState.Confirmed
def is_deleted(self):
"""Returns True if this track is dead and should be deleted."""
return self.state == TrackState.Deleted
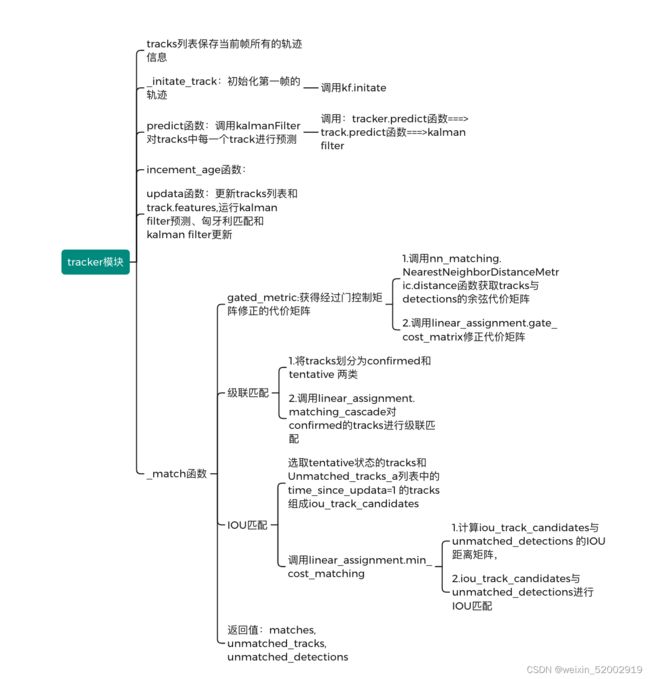
4.6 tracker 模块
# vim: expandtab:ts=4:sw=4
from __future__ import absolute_import
import numpy as np
from . import kalman_filter
from . import linear_assignment
from . import iou_matching
from .track import Track
class Tracker:
"""
This is the multi-target tracker.
Parameters
----------
metric : nn_matching.NearestNeighborDistanceMetric
A distance metric for measurement-to-track association.
max_age : int
Maximum number of missed misses before a track is deleted.
n_init : int
Number of consecutive detections before the track is confirmed. The
track state is set to `Deleted` if a miss occurs within the first
`n_init` frames.
Attributes
----------
metric : nn_matching.NearestNeighborDistanceMetric
The distance metric used for measurement to track association.
max_age : int
Maximum number of missed misses before a track is deleted.
n_init : int
Number of frames that a track remains in initialization phase.
kf : kalman_filter.KalmanFilter
A Kalman filter to filter target trajectories in image space.
tracks : List[Track]
The list of active tracks at the current time step.
"""
def __init__(self, metric, max_iou_distance=0.7, max_age=70, n_init=3):
self.metric = metric
self.max_iou_distance = max_iou_distance
self.max_age = max_age
self.n_init = n_init
self.kf = kalman_filter.KalmanFilter()
self.tracks = [] #
self._next_id = 1
def predict(self):
"""Propagate track state distributions one time step forward.
This function should be called once every time step, before `update`.
"""
for track in self.tracks:
track.predict(self.kf)
def increment_ages(self):
for track in self.tracks:
track.increment_age()
track.mark_missed()
def update(self, detections):
"""Perform measurement update and track management.
Parameters
----------
detections : List[deep_sort.detection.Detection]
A list of detections at the current time step.
"""
# Run matching cascade.
matches, unmatched_tracks, unmatched_detections = \
self._match(detections)
# Update track set.
for track_idx, detection_idx in matches:
self.tracks[track_idx].update(
self.kf, detections[detection_idx])
for track_idx in unmatched_tracks:
self.tracks[track_idx].mark_missed()
for detection_idx in unmatched_detections:
self._initiate_track(detections[detection_idx])
self.tracks = [t for t in self.tracks if not t.is_deleted()]
# Update distance metric.
active_targets = [t.track_id for t in self.tracks if t.is_confirmed()]
features, targets = [], []
for track in self.tracks:
if not track.is_confirmed():
continue
features += track.features
targets += [track.track_id for _ in track.features]
track.features = []
self.metric.partial_fit(
np.asarray(features), np.asarray(targets), active_targets)
def _match(self, detections):
def gated_metric(tracks, dets, track_indices, detection_indices):
features = np.array([dets[i].feature for i in detection_indices])
targets = np.array([tracks[i].track_id for i in track_indices])
cost_matrix = self.metric.distance(features, targets)
cost_matrix = linear_assignment.gate_cost_matrix(
self.kf, cost_matrix, tracks, dets, track_indices,
detection_indices)
return cost_matrix
# Split track set into confirmed and unconfirmed tracks.
confirmed_tracks = [
i for i, t in enumerate(self.tracks) if t.is_confirmed()]
unconfirmed_tracks = [
i for i, t in enumerate(self.tracks) if not t.is_confirmed()]
# Associate confirmed tracks using appearance features.
matches_a, unmatched_tracks_a, unmatched_detections = \
linear_assignment.matching_cascade(
gated_metric, self.metric.matching_threshold, self.max_age,
self.tracks, detections, confirmed_tracks)
# Associate remaining tracks together with unconfirmed tracks using IOU.
iou_track_candidates = unconfirmed_tracks + [
k for k in unmatched_tracks_a if
self.tracks[k].time_since_update == 1]
unmatched_tracks_a = [
k for k in unmatched_tracks_a if
self.tracks[k].time_since_update != 1]
matches_b, unmatched_tracks_b, unmatched_detections = \
linear_assignment.min_cost_matching(
iou_matching.iou_cost, self.max_iou_distance, self.tracks,
detections, iou_track_candidates, unmatched_detections)
matches = matches_a + matches_b
unmatched_tracks = list(set(unmatched_tracks_a + unmatched_tracks_b))
return matches, unmatched_tracks, unmatched_detections
def _initiate_track(self, detection):
mean, covariance = self.kf.initiate(detection.to_xyah())
self.tracks.append(Track(
mean, covariance, self._next_id, self.n_init, self.max_age,
detection.feature))
self._next_id += 1
Feature_extractor模块
import torch
import torchvision.transforms as transforms
import numpy as np
import cv2
import logging
from .model import Net
class Extractor(object):
def __init__(self, model_path, use_cuda=True):
self.net = Net(reid=True)
self.device = "cuda" if torch.cuda.is_available() and use_cuda else "cpu"
state_dict = torch.load(model_path, map_location=lambda storage, loc: storage)[
'net_dict']
self.net.load_state_dict(state_dict)
logger = logging.getLogger("root.tracker")
logger.info("Loading weights from {}... Done!".format(model_path))
self.net.to(self.device)
self.size = (64, 128)
self.norm = transforms.Compose([
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225]),
])
def _preprocess(self, im_crops):
"""
TODO:
1. to float with scale from 0 to 1
2. resize to (64, 128) as Market1501 dataset did
3. concatenate to a numpy array
3. to torch Tensor
4. normalize
"""
def _resize(im, size):
return cv2.resize(im.astype(np.float32)/255., size)
im_batch = torch.cat([self.norm(_resize(im, self.size)).unsqueeze(
0) for im in im_crops], dim=0).float()
return im_batch
def __call__(self, im_crops):
im_batch = self._preprocess(im_crops)
with torch.no_grad():
im_batch = im_batch.to(self.device)
features = self.net(im_batch)
return features.cpu().numpy()
if __name__ == '__main__':
img = cv2.imread("demo.jpg")[:, :, (2, 1, 0)]
extr = Extractor("checkpoint/ckpt.t7")
feature = extr(img)
print(feature.shape)
deep_sort 模块
import numpy as np
import torch
from .deep.feature_extractor import Extractor
from .sort.nn_matching import NearestNeighborDistanceMetric
from .sort.preprocessing import non_max_suppression
from .sort.detection import Detection
from .sort.tracker import Tracker
__all__ = ['DeepSort']
class DeepSort(object):
def __init__(self, model_path, max_dist=0.2, min_confidence=0.3, nms_max_overlap=1.0, max_iou_distance=0.7, max_age=70, n_init=3, nn_budget=100, use_cuda=True):
self.min_confidence = min_confidence
self.nms_max_overlap = nms_max_overlap
self.extractor = Extractor(model_path, use_cuda=use_cuda)
max_cosine_distance = max_dist
metric = NearestNeighborDistanceMetric(
"cosine", max_cosine_distance, nn_budget)
self.tracker = Tracker(
metric, max_iou_distance=max_iou_distance, max_age=max_age, n_init=n_init)
def update(self, bbox_xywh, confidences, ori_img):
self.height, self.width = ori_img.shape[:2]
# generate detections
features = self._get_features(bbox_xywh, ori_img)
bbox_tlwh = self._xywh_to_tlwh(bbox_xywh)
detections = [Detection(bbox_tlwh[i], conf, features[i]) for i, conf in enumerate(
confidences) if conf > self.min_confidence]
# run on non-maximum supression(非极大值抑制)
boxes = np.array([d.tlwh for d in detections])
scores = np.array([d.confidence for d in detections])
indices = non_max_suppression(boxes, self.nms_max_overlap, scores)
detections = [detections[i] for i in indices]
# update tracker
self.tracker.predict()
self.tracker.update(detections)
# output bbox identities
outputs = []
for track in self.tracker.tracks:
if not track.is_confirmed() or track.time_since_update > 1:
continue
box = track.to_tlwh()
x1, y1, x2, y2 = self._tlwh_to_xyxy(box)
track_id = track.track_id
outputs.append(np.array([x1, y1, x2, y2, track_id], dtype=np.int))
if len(outputs) > 0:
outputs = np.stack(outputs, axis=0)
return outputs
"""
TODO:
Convert bbox from xc_yc_w_h to xtl_ytl_w_h
Thanks [email protected] for reporting this bug!
"""
@staticmethod
def _xywh_to_tlwh(bbox_xywh):
if isinstance(bbox_xywh, np.ndarray):
bbox_tlwh = bbox_xywh.copy()
elif isinstance(bbox_xywh, torch.Tensor):
bbox_tlwh = bbox_xywh.clone()
bbox_tlwh[:, 0] = bbox_xywh[:, 0] - bbox_xywh[:, 2] / 2.
bbox_tlwh[:, 1] = bbox_xywh[:, 1] - bbox_xywh[:, 3] / 2.
return bbox_tlwh
def _xywh_to_xyxy(self, bbox_xywh):
x, y, w, h = bbox_xywh
x1 = max(int(x - w / 2), 0)
x2 = min(int(x + w / 2), self.width - 1)
y1 = max(int(y - h / 2), 0)
y2 = min(int(y + h / 2), self.height - 1)
return x1, y1, x2, y2
def _tlwh_to_xyxy(self, bbox_tlwh):
"""
TODO:
Convert bbox from xtl_ytl_w_h to xc_yc_w_h
Thanks [email protected] for reporting this bug!
"""
x, y, w, h = bbox_tlwh
x1 = max(int(x), 0)
x2 = min(int(x+w), self.width - 1)
y1 = max(int(y), 0)
y2 = min(int(y+h), self.height - 1)
return x1, y1, x2, y2
def increment_ages(self):
self.tracker.increment_ages()
def _xyxy_to_tlwh(self, bbox_xyxy):
x1, y1, x2, y2 = bbox_xyxy
t = x1
l = y1
w = int(x2 - x1)
h = int(y2 - y1)
return t, l, w, h
def _get_features(self, bbox_xywh, ori_img):
im_crops = []
for box in bbox_xywh:
x1, y1, x2, y2 = self._xywh_to_xyxy(box)
im = ori_img[y1:y2, x1:x2]
im_crops.append(im)
if im_crops:
features = self.extractor(im_crops)
else:
features = np.array([])
return features