人工晶状体计算——人工智能算法(R语言)
人工晶状体计算——人工智能算法(R语言)
1. 准备数据
2. 建立模型
2.1 方法1
2.2 方法2
1. 准备数据
准备数据Data.xlsx,示例如图
| Age | AL | ACD | K1 | K2 | WTW | LT | Ref | IOLPower |
| 68.00 | 22.68 | 2.21 | 42.44 | 42.75 | 11.20 | 4.82 | -1.13 | 26.50 |
| 62.00 | 23.79 | 3.42 | 43.93 | 45.51 | 11.70 | 4.49 | -0.11 | 19.50 |
| 62.00 | 23.82 | 3.12 | 44.08 | 44.53 | 12.00 | 4.90 | -0.86 | 21.00 |
| 68.00 | 22.56 | 2.30 | 42.65 | 43.13 | 11.00 | 4.65 | -0.37 | 25.50 |
| 73.00 | 23.50 | 2.38 | 43.43 | 43.84 | 11.40 | 4.85 | -0.26 | 21.50 |
| 74.00 | 23.09 | 2.91 | 43.86 | 44.74 | 11.90 | 4.45 | -0.53 | 23.00 |
| 73.00 | 24.61 | 3.57 | 41.70 | 42.23 | 11.90 | 3.49 | -0.80 | 21.00 |
| 64.00 | 22.47 | 2.89 | 46.63 | 48.01 | 11.10 | 4.41 | -0.52 | 21.00 |
| 63.00 | 21.35 | 2.67 | 43.89 | 44.53 | 12.20 | 4.68 | -0.58 | 29.50 |
2. 建立模型
2.1 方法1
直接建立Age,AL,ACD,K1,K2,WTW,LT,Ref与IOLPower之间的关系
将使用多种人工智能模型,最后将表现最优的三种叠加在一起。
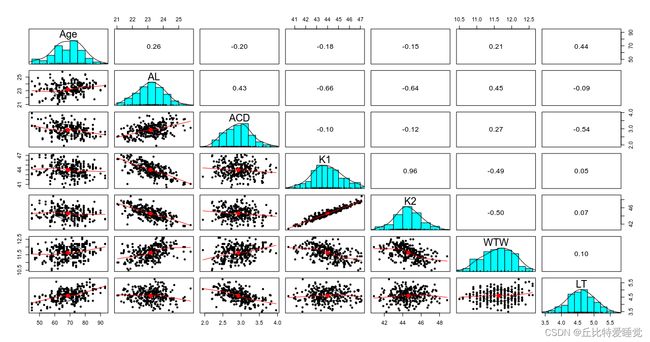
开始先看一下各自变量之间的相关性:
可以看到自变量之间还是有一定相关性的,比如LT(Lens Thickness)与Age正相关,ACD与AL正相关等。
rm(list=ls())
# load libraries
library(caret)
library(corrplot)
library(tidyverse)
library(xlsx)
# Split out validation dataset
set.seed(123)
df <- read.xlsx("./Data.xlsx",sheetName ="Sheet1",stringsAsFactors = FALSE)
str(df)
names(df)<-c("Age","AL","ACD","K1","K2","WTW","LT","IOLPower","Ref")
cor(df[c("Age","AL","ACD","K1","K2","WTW","LT")])
pairs(df[c("Age","AL","ACD","K1","K2","WTW","LT")])
# more informative scatterplot matrix
library(psych)
pairs.panels(df[c("Age","AL","ACD","K1","K2","WTW","LT")])
train.idx <- sample(1:nrow(df), 0.7*nrow(df))
train.df <- as_tibble(df[train.idx,])
test.df <- as_tibble(df[-train.idx,])
train_x = train.df %>% select(-'IOLPower')
train_y = train.df[,'IOLPower']
test_x = test.df %>% select(-'IOLPower')
test_y = test.df[,'IOLPower']
# Run algorithms using 10-fold cross validation
control <- trainControl(method="repeatedcv", number=10, repeats=3, search="random")
metric <- "RMSE"
# lm
set.seed(123)
fit_lm <- train(IOLPower ~ Age+AL+ACD+K1+K2+WTW+LT+Ref, data=train.df, method="lm",
metric=metric, preProc=c("center", "scale"), tuneLength=15, trControl=control)
fit_lm
# SVM
set.seed(123)
fit_svm <- train(IOLPower ~ Age+AL+ACD+K1+K2+WTW+LT+Ref,data=train.df,
method="svmRadial", metric=metric, preProc=c("center", "scale"), tuneLength=15,
trControl=control)
fit_svm
# ANNs
fit_nnet <- train(IOLPower ~ Age+AL+ACD+K1+K2+WTW+LT+Ref,data=train.df, method="nnet",
metric=metric, preProc=c("center", "scale"), tuneLength=15, trControl=control)
fit_nnet
if(FALSE)
{
pred_Power <-predict(fit_nnet,test_x)
print(pred_Power)
err<- test_y$IOLPower-pred_Power
print(err)
plot(err)
}
# random forest
set.seed(123)
fit_rf <- train(IOLPower ~ Age+AL+ACD+K1+K2+WTW+LT+Ref, data = train.df, method="rf",
metric=metric, preProc=c("center", "scale"), tuneLength=15, trControl=control)
fit_rf
# CART
set.seed(123)
fit_cart <- train(IOLPower ~ Age+AL+ACD+K1+K2+WTW+LT+Ref,data=train.df, method="rpart",
metric=metric,preProc=c("center", "scale"),tuneLength=15, trControl=control)
fit_cart
# kNN
set.seed(123)
fit_knn <- train(IOLPower ~ Age+AL+ACD+K1+K2+WTW+LT+Ref,data=train.df, method="knn",
metric=metric, preProc=c("center", "scale"),tuneLength=15, trControl=control)
fit_knn
#MARS
set.seed(123)
fit_MARS<- train(IOLPower ~ Age+AL+ACD+K1+K2+WTW+LT+Ref,data=train.df,
method="earth", metric=metric, preProc=c("center", "scale"), tuneLength=15, trControl=control)
fit_MARS
# GBM
set.seed(123)
fit_gbm <- train(IOLPower ~ Age+AL+ACD+K1+K2+WTW+LT+Ref, data = train.df, method =
"gbm",metric=metric,preProc=c("center", "scale"), tuneLength=15, trControl=control)
fit_gbm
#cubist
set.seed(123)
trControl=control
fit_cubist<- train(IOLPower ~ Age+AL+ACD+K1+K2+WTW+LT+Ref, data = train.df, method =
"cubist",metric=metric,preProc=c("center", "scale"), tuneLength=15, trControl=control)
fit_cubist
# Compare algorithms
results <- resamples(list(LM = fit_lm, CART = fit_cart, KNN= fit_knn, RF = fit_rf, SVR = fit_svm,
GBM = fit_gbm, MARS= fit_MARS, Cubist = fit_cubist,ANNs=fit_nnet))
summary(results)
dotplot(results)
# Stacking
library(caretEnsemble)
set.seed(123)
allModels <- caretList(IOLPower ~ Age+AL+ACD+K1+K2+WTW+LT+Ref,data=train.df,trControl
= trainControl("repeatedcv", number = 10, repeats = 3, search="random",verboseIter = T),metric =
"RMSE", tuneList = list(cubist = caretModelSpec(method = "cubist", tuneLength = 15, preProc =
c("center","scale")),svmRadial = caretModelSpec(method = "svmRadial", tuneLength = 15, preProc
= c("center","scale")),Earth = caretModelSpec(method = "earth", tuneLength = 15, preProc =
c("center","scale"))))
bwplot(resamples(allModels), metric = "RMSE")
summary(allModels)
# Stack with lm
set.seed(123)
stack <- caretStack(allModels, metric = "RMSE", method = "lm", tuneLength = 10,trControl =
trainControl("repeatedcv", number = 5, repeats = 3, verboseIter = T))
summary(stack)
print(stack)
if(TRUE){
ggplot(varImp(fit_lm))
pred_Power <-predict(stack,test_x)
print(pred_Power)
plot(pred_Power)
pred_err <- test_y$IOLPower - pred_Power
print(pred_err)
summary(pred_err)
plot(pred_err)
}运行结果误差还是很小的。
2.2 方法2
结合光学公式与人工智能(此法源自中山的一篇论文)
光学人工晶体计算公式如下
Power =1336 / (AL- ELP) - (1336/(1336 /(1000 /(1000 /Ref-12)+K)-ELP))临床实践中最难的就是预测ELP(Effective Lens Position,有效人工晶状体位置,个人觉得"等效人工晶状体位置"这个翻译可能更好),以下代码中Ratio指
Ratio =(ELP-ACD)/LT下面代码的各个模型都是为了预测这个Ratio,最后求得ELP,然后代入上述光学计算公式中得到最后的人工晶状体度数。
if (TRUE){
rm(list=ls())
library(xlsx)
data <- read.xlsx("./Data.xlsx",sheetName ="Sheet1",stringsAsFactors = FALSE)
f<-function(ELP,AL,K,Ref,P)
{1336 / (AL- ELP) - (1336/(1336 /(1000 /(1000 /Ref-12)+K)-ELP))-P}
root<-c()
for(i in 1:nrow(data)){
root[i]<-uniroot(f,c(0,20),AL=data$AL[i],K=(data$K1[i]+data$K2[i])/2,R=data$Ref[i],P=data$IOLPower[i])$root
}
root
data$ELP <-root
Ratio <- (data$ELP-data$ACD)/data$LT
Ratio
data$Ratio<-Ratio
PowerCalc<-function(ELP,AL,K,Ref)
{1336 / (AL- ELP) - (1336/(1336 /(1000 /(1000 /Ref-12)+K)-ELP))}
P<-PowerCalc(data$ELP,data$AL,(data$K1+data$K2)/2,data$Ref)
P
options(scipen = 100) #小数点后100位不使用科学计数法
err <- data$IOLPower - P
summary(err)
plot(err)
}
library(caret)
library(corrplot)
library(tidyverse)
library(xlsx)
# Split out validation dataset
set.seed(123)
cor(data[c("Age","AL","ACD","K1","K2","WTW","LT")])
pairs(data[c("Age","AL","ACD","K1","K2","WTW","LT")])
# more informative scatterplot matrix
library(psych)
pairs.panels(data[c("Age","AL","ACD","K1","K2","WTW","LT")])
train.idx <- sample(1:nrow(data), 0.7*nrow(data))
train.data <- as_tibble(data[train.idx,])
test.data <- as_tibble(data[-train.idx,])
train_x = train.data %>% select(-'Ratio')
train_y = train.data[,'Ratio']
test_x = test.data %>% select(-'Ratio')
test_y = test.data[,'Ratio']
# Run algorithms using 10-fold cross validation
control <- trainControl(method="repeatedcv", number=10, repeats=3, search="random")
metric <- "RMSE"
# lm
set.seed(123)
fit_lm <- train(Ratio ~ Age+AL+ACD+K1+K2+WTW+LT+Ref, data=train.data, method="lm",
metric=metric, preProc=c("center", "scale"), tuneLength=15, trControl=control)
fit_lm
# SVM
set.seed(123)
fit_svm <- train(Ratio ~ Age+AL+ACD+K1+K2+WTW+LT+Ref,data=train.data,
method="svmRadial", metric=metric, preProc=c("center", "scale"), tuneLength=15,
trControl=control)
fit_svm
plot(fit_svm)
# ANNs
fit_nnet <- train(Ratio ~ Age+AL+ACD+K1+K2+WTW+LT+Ref,data=train.data, method="nnet",
metric=metric, preProc=c("center", "scale"), tuneLength=15, trControl=control)
fit_nnet
if(TRUE)
{
pred <-predict(fit_nnet,test_x)
print(pred)
err<- test_y$Ratio-pred
print(err)
plot(err)
}
# random forest
set.seed(123)
fit_rf <- train(Ratio ~ Age+AL+ACD+K1+K2+WTW+LT+Ref, data = train.data, method="rf",
metric=metric, preProc=c("center", "scale"), tuneLength=15, trControl=control)
fit_rf
# CART
set.seed(123)
fit_cart <- train(Ratio ~ Age+AL+ACD+K1+K2+WTW+LT+Ref,data=train.data, method="rpart",
metric=metric,preProc=c("center", "scale"),tuneLength=15, trControl=control)
fit_cart
# kNN
set.seed(123)
fit_knn <- train(Ratio ~ Age+AL+ACD+K1+K2+WTW+LT+Ref,data=train.data, method="knn",
metric=metric, preProc=c("center", "scale"),tuneLength=15, trControl=control)
fit_knn
#MARS
set.seed(123)
fit_MARS<- train(Ratio ~ Age+AL+ACD+K1+K2+WTW+LT+Ref,data=train.data,
method="earth", metric=metric, preProc=c("center", "scale"), tuneLength=15, trControl=control)
fit_MARS
# GBM
set.seed(123)
fit_gbm <- train(Ratio ~ Age+AL+ACD+K1+K2+WTW+LT+Ref, data = train.data, method =
"gbm",metric=metric,preProc=c("center", "scale"), tuneLength=15, trControl=control)
fit_gbm
#cubist
set.seed(123)
trControl=control
fit_cubist<- train(Ratio ~ Age+AL+ACD+K1+K2+WTW+LT+Ref, data = train.data, method =
"cubist",metric=metric,preProc=c("center", "scale"), tuneLength=15, trControl=control)
fit_cubist
# Compare algorithms
results <- resamples(list(LM = fit_lm, CART = fit_cart, KNN= fit_knn, RF = fit_rf, SVR = fit_svm,
GBM = fit_gbm, MARS= fit_MARS, Cubist = fit_cubist,ANNs=fit_nnet))
summary(results)
dotplot(results)
# Stacking
library(caretEnsemble)
set.seed(123)
allModels <- caretList(Ratio ~ Age+AL+ACD+K1+K2+WTW+LT+Ref,data=train.data,trControl
= trainControl("repeatedcv", number = 10, repeats = 3, search="random",verboseIter = T),metric =
"RMSE", tuneList = list(cubist = caretModelSpec(method = "cubist", tuneLength = 15, preProc =
c("center","scale")),svmRadial = caretModelSpec(method = "svmRadial", tuneLength = 15, preProc
= c("center","scale")),Earth = caretModelSpec(method = "earth", tuneLength = 15, preProc =
c("center","scale"))))
bwplot(resamples(allModels), metric = "RMSE")
summary(allModels)
# Stack with lm
set.seed(123)
stack <- caretStack(allModels, metric = "RMSE", method = "lm", tuneLength = 10,trControl =
trainControl("repeatedcv", number = 5, repeats = 3, verboseIter = T))
summary(stack)
print(stack)
if(TRUE){
ggplot(varImp(fit_nnet))
pred_Ratio <-predict(stack,test_x)
print(pred_Ratio)
plot(pred_Ratio)
pred_err1 <- test_y$Ratio - pred_Ratio
print(pred_err1)
summary(pred_err1)
plot(pred_err1)
ELPCalc<-function(Ratio,LT,ACD){Ratio*LT+ACD}
ELP = ELPCalc(pred_Ratio,test_x$LT,test_x$ACD)
Pred_Power<-PowerCalc(ELP,test_x$AL,(test_x$K1+test_x$K2)/2,test_x$Ref)
Pred_err2<-test_x$IOLPower - Pred_Power
summary(Pred_err2)
plot(Pred_err2)
}复现论文也算勉强成功,但是样本量太少,暂无法比较两种方法的优劣。