2022讯飞——糖尿病遗传风险检测挑战赛解决方案
目录:
- 0. 赛事背景
- 1. 读取数据
- 2. 数据探索及预处理
-
- 2.1 缺失值
- 2.2 分析字段类型
- 2.3 计算字段相关性
- 3. 特征工程
-
- 3.1 特征构造
- 4. 模型训练
-
- 4.1 LightGBM (0.96206)
- 4.2 随机森林(0.96324)
- 4.3 XGBoost (0.95981)
- 4.4 CatBoost(0.95854)
- 4.5 AdaBoost(0.96098)
- 4.6 集成模型(0.95971)
- 4.7 Stacking(0.96577)
- 4.8 归一化数据,pytorch神经网络
- 4.9 SVM
- 4.10 sklearn神经网络
- 5. 总结思考
0. 赛事背景
截至2022年,中国糖尿病患者近1.3亿。中国糖尿病患病原因受生活方式、老龄化、城市化、家族遗传等多种因素影响。同时,糖尿病患者趋向年轻化。
糖尿病可导致心血管、肾脏、脑血管并发症的发生。因此,准确诊断出患有糖尿病个体具有非常重要的临床意义。糖尿病早期遗传风险预测将有助于预防糖尿病的发生。
根据《中国2型糖尿病防治指南(2017年版)》,糖尿病的诊断标准是具有典型糖尿病症状(烦渴多饮、多尿、多食、不明原因的体重下降)且随机静脉血浆葡萄糖≥11.1mmol/L或空腹静脉血浆葡萄糖≥7.0mmol/L或口服葡萄糖耐量试验(OGTT)负荷后2h血浆葡萄糖≥11.1mmol/L。
在这次比赛中,您需要通过训练数据集构建糖尿病遗传风险预测模型,然后预测出测试数据集中个体是否患有糖尿病,和我们一起帮助糖尿病患者解决这“甜蜜的烦恼”。
-
训练集说明
训练集(比赛训练集.csv)一共有5070条数据,用于构建您的预测模型(您可能需要先进行数据分析)。数据的字段有编号、性别、出生年份、体重指数、糖尿病家族史、舒张压、口服耐糖量测试、胰岛素释放实验、肱三头肌皮褶厚度、患有糖尿病标识(最后一列),您也可以通过特征工程技术构建新的特征。 -
测试集说明
测试集(比赛测试集.csv)一共有1000条数据,用于验证预测模型的性能。数据的字段有编号、性别、出生年份、体重指数、糖尿病家族史、舒张压、口服耐糖量测试、胰岛素释放实验、肱三头肌皮褶厚度。 -
评估指标
对于提交的结果,系统会采用二分类任务中的F1-score指标进行评价,F1-score越大说明预测模型性能越好。
本质为二分类预测,不是很难,注意做好特征工程,提升数据质量。
比赛详情见官网。
1. 读取数据
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import lightgbm
train_data = pd.read_csv('比赛训练集.csv',encoding='gbk')
test_data = pd.read_csv('比赛测试集.csv',encoding='gbk')
train_data.describe()
| 编号 | 性别 | 出生年份 | 体重指数 | 舒张压 | 口服耐糖量测试 | 胰岛素释放实验 | 肱三头肌皮褶厚度 | 患有糖尿病标识 | |
|---|---|---|---|---|---|---|---|---|---|
| count | 5070.000000 | 5070.000000 | 5070.000000 | 5070.000000 | 4823.000000 | 5070.000000 | 5070.000000 | 5070.000000 | 5070.000000 |
| mean | 2535.500000 | 0.456805 | 1986.869231 | 37.986785 | 89.423595 | 5.612839 | 4.114321 | 6.994371 | 0.381854 |
| std | 1463.727263 | 0.498180 | 8.919737 | 11.447095 | 9.266992 | 2.257649 | 8.726001 | 13.651442 | 0.485889 |
| min | 1.000000 | 0.000000 | 1943.000000 | 0.000000 | 30.000000 | -1.000000 | 0.000000 | 0.000000 | 0.000000 |
| 25% | 1268.250000 | 0.000000 | 1980.000000 | 28.400000 | 85.000000 | 4.314000 | 0.000000 | 0.000000 | 0.000000 |
| 50% | 2535.500000 | 0.000000 | 1987.000000 | 36.550000 | 89.000000 | 5.760000 | 0.000000 | 0.000000 | 0.000000 |
| 75% | 3802.750000 | 1.000000 | 1995.000000 | 47.600000 | 96.000000 | 7.193000 | 7.100000 | 4.120000 | 1.000000 |
| max | 5070.000000 | 1.000000 | 2009.000000 | 65.900000 | 126.000000 | 10.839000 | 108.960000 | 45.000000 | 1.000000 |
患有糖尿病标识的均值为0.38,说明未患病:患病 ≈ 6 : 4 \approx 6 : 4 ≈6:4,数据还算较为平衡。
test_data.describe()
| 编号 | 性别 | 出生年份 | 体重指数 | 舒张压 | 口服耐糖量测试 | 胰岛素释放实验 | 肱三头肌皮褶厚度 | |
|---|---|---|---|---|---|---|---|---|
| count | 1000.000000 | 1000.000000 | 1000.000000 | 1000.000000 | 951.000000 | 1000.000000 | 1000.000000 | 1000.000000 |
| mean | 500.500000 | 0.481000 | 1986.386000 | 39.439000 | 89.638275 | 5.872314 | 4.102700 | 7.064240 |
| std | 288.819436 | 0.499889 | 8.816163 | 11.284861 | 9.379124 | 1.930880 | 8.594005 | 13.900938 |
| min | 1.000000 | 0.000000 | 1958.000000 | 0.000000 | 28.000000 | -1.000000 | 0.000000 | 0.000000 |
| 25% | 250.750000 | 0.000000 | 1979.000000 | 29.975000 | 85.000000 | 4.516000 | 0.000000 | 0.000000 |
| 50% | 500.500000 | 0.000000 | 1987.000000 | 38.900000 | 89.000000 | 5.851500 | 0.000000 | 0.000000 |
| 75% | 750.250000 | 1.000000 | 1994.000000 | 48.950000 | 96.000000 | 7.465000 | 7.202500 | 3.820000 |
| max | 1000.000000 | 1.000000 | 2003.000000 | 60.000000 | 112.000000 | 10.613000 | 123.890000 | 44.900000 |
2. 数据探索及预处理
2.1 缺失值
统计每个字段的缺失比例,并进行填充。可以看到舒张压指标的缺失值较多,用字段均值将其填充。
print('训练集各字段缺失比例:')
print(train_data.isnull().mean(0))
print('\n测试集各字段缺失比例:')
print(test_data.isnull().mean(0))
# 用均值填充缺失值
train_data['舒张压'] = train_data['舒张压'].fillna(train_data['舒张压'].mean())
test_data['舒张压'] = test_data['舒张压'].fillna(test_data['舒张压'].mean())
训练集各字段缺失比例:
编号 0.000000
性别 0.000000
出生年份 0.000000
体重指数 0.000000
糖尿病家族史 0.000000
舒张压 0.048718
口服耐糖量测试 0.000000
胰岛素释放实验 0.000000
肱三头肌皮褶厚度 0.000000
患有糖尿病标识 0.000000
dtype: float64
测试集各字段缺失比例:
编号 0.000
性别 0.000
出生年份 0.000
体重指数 0.000
糖尿病家族史 0.000
舒张压 0.049
口服耐糖量测试 0.000
胰岛素释放实验 0.000
肱三头肌皮褶厚度 0.000
dtype: float64
2.2 分析字段类型
print(train_data.columns)
train_data.describe()
Index(['编号', '性别', '出生年份', '体重指数', '糖尿病家族史', '舒张压', '口服耐糖量测试', '胰岛素释放实验',
'肱三头肌皮褶厚度', '患有糖尿病标识'],
dtype='object')
| 编号 | 性别 | 出生年份 | 体重指数 | 舒张压 | 口服耐糖量测试 | 胰岛素释放实验 | 肱三头肌皮褶厚度 | 患有糖尿病标识 | |
|---|---|---|---|---|---|---|---|---|---|
| count | 5070.000000 | 5070.000000 | 5070.000000 | 5070.000000 | 5070.000000 | 5070.000000 | 5070.000000 | 5070.000000 | 5070.000000 |
| mean | 2535.500000 | 0.456805 | 1986.869231 | 37.986785 | 89.423595 | 5.612839 | 4.114321 | 6.994371 | 0.381854 |
| std | 1463.727263 | 0.498180 | 8.919737 | 11.447095 | 9.038394 | 2.257649 | 8.726001 | 13.651442 | 0.485889 |
| min | 1.000000 | 0.000000 | 1943.000000 | 0.000000 | 30.000000 | -1.000000 | 0.000000 | 0.000000 | 0.000000 |
| 25% | 1268.250000 | 0.000000 | 1980.000000 | 28.400000 | 85.000000 | 4.314000 | 0.000000 | 0.000000 | 0.000000 |
| 50% | 2535.500000 | 0.000000 | 1987.000000 | 36.550000 | 89.000000 | 5.760000 | 0.000000 | 0.000000 | 0.000000 |
| 75% | 3802.750000 | 1.000000 | 1995.000000 | 47.600000 | 95.000000 | 7.193000 | 7.100000 | 4.120000 | 1.000000 |
| max | 5070.000000 | 1.000000 | 2009.000000 | 65.900000 | 126.000000 | 10.839000 | 108.960000 | 45.000000 | 1.000000 |
编号与是否患病没关系,删除;
性别为类别变量,只有0,1,不再需要进行编码;
糖尿病家族病史为文本型变量,需要转化为数值变量;
其他均为数值型变量,可以暂时不变。
train_data = train_data.drop(['编号'], axis=1)
test_data = test_data.drop(['编号'], axis=1)
2.3 计算字段相关性
查看各字段之间的相关性,防止多重共线性。
Ref:
Python绘制相关性热力图
train_corr = train_data.drop('糖尿病家族史',axis=1).corr()
import seaborn as sns
plt.subplots(figsize=(9,9),dpi=80,facecolor='w') # 设置画布大小,分辨率,和底色
plt.rcParams['font.sans-serif'] = ['SimHei'] # 黑体
plt.rcParams['axes.unicode_minus'] = False # 解决无法显示符号的问题
sns.set(font='SimHei', font_scale=0.8) # 解决Seaborn中文显示问题
#annot为热力图上显示数据;fmt='.2g'为数据保留两位有效数字,square呈现正方形,vmax最大值为1
fig=sns.heatmap(train_corr,annot=True, vmax=1, square=True, cmap="Blues", fmt='.2g')
#保存图片
fig.get_figure().savefig('train_corr.png',bbox_inches='tight',transparent=True)
#bbox_inches让图片显示完整,transparent=True让图片背景透明
这里中文显示出了点问题,但是可以看到各特征变量与是否患病没有显著线性关系(但可能有非线性关系),各特征变量之间也不存在多重共线性,可以继续下一步操作。
3. 特征工程
这一步至关重要,主要是有两个目的:
- 特征构造:尝试构建有价值的新变量;
- 特征筛选:删除对因变量影响不大的冗余变量。
由于这里的特征也不是很多,就不做筛选了。
3.1 特征构造
可以用统计指标,已有知识、经验构造新的变量,具体到这个问题上可以有BMI指数、舒张压范围、年龄等。
特征构造方法:
- 特征的统计指标;
- 特征之间的四则运算;
- 交叉特征;
- 分解类别特征。如将三个颜色分解为“知道颜色”和“不知道颜色”。
- 特征分箱。将数值型特征变量按段划分,得到类别型特征。
- 重构特征。单位转换、整数部分与小数部分分离等。
- 根据已有经验构造新的特征变量,比如xx因子。
Ref:
[1] 深度了解特征工程
# 将出生年份换算成年龄
train_data['年龄']=2022-train_data['出生年份'] #换成年龄
test_data['年龄']=2022-test_data['出生年份']
train_data = train_data.drop('出生年份', axis=1)
test_data = test_data.drop('出生年份', axis=1)
# 家族史转换, 方法一,label编码
from sklearn.preprocessing import OneHotEncoder, LabelEncoder
def FHOD(a):
if a=='无记录':
return 0
elif a=='叔叔或者姑姑有一方患有糖尿病' or a=='叔叔或姑姑有一方患有糖尿病':
return 1
else:
return 2
train_data['糖尿病家族史'] = train_data['糖尿病家族史'].apply(FHOD)
test_data['糖尿病家族史'] = test_data['糖尿病家族史'].apply(FHOD)
# history = train_data['糖尿病家族史']
# print(set(history))
# history.loc[history=='叔叔或姑姑有一方患有糖尿病'] = '叔叔或者姑姑有一方患有糖尿病'
# le = LabelEncoder()
# h = le.fit_transform(history)
# 方法二,onehot 编码
# def onehot_transform(data):
# # 将家族史的文本型变量转换为onehot编码。
# onehot = OneHotEncoder()
# data.loc[data['糖尿病家族史']=='叔叔或姑姑有一方患有糖尿病', '糖尿病家族史'] = '叔叔或者姑姑有一方患有糖尿病'
# data_onehot = pd.DataFrame(onehot.fit_transform(data[['糖尿病家族史']]).toarray(),
# columns=onehot.get_feature_names(['糖尿病家族史']), dtype='int32')
# return data_onehot
# data_train_history = onehot_transform(train_data)
# data_test_history = onehot_transform(test_data)
def BMI(a):
"""
人体的成人体重指数正常值是在18.5-24之间
低于18.5是体重指数过轻
在24-27之间是体重超重
27以上考虑是肥胖
高于32了就是非常的肥胖。
"""
if a<18.5:
return 0
elif 18.5<=a<=24:
return 1
elif 24<a<=27:
return 2
elif 27<a<=32:
return 3
else:
return 4
train_data['BMI']=train_data['体重指数'].apply(BMI)
test_data['BMI']=test_data['体重指数'].apply(BMI)
# 转换舒张压为类别型变量
def DBP(a):
# 舒张压范围为60-90
if a<60:
return 0
elif 60<=a<=90:
return 1
elif a>90:
return 2
else:
return a
train_data['DBP'] = train_data['舒张压'].apply(DBP)
test_data['DBP'] = test_data['舒张压'].apply(DBP)
X_train = train_data.drop('患有糖尿病标识', axis=1)
Y_train = train_data['患有糖尿病标识']
X_train['年龄'] = X_train['年龄'].astype(float)
X_test = test_data
train_data.describe()
| 性别 | 体重指数 | 糖尿病家族史 | 舒张压 | 口服耐糖量测试 | 胰岛素释放实验 | 肱三头肌皮褶厚度 | 患有糖尿病标识 | 年龄 | BMI | DBP | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 5070.000000 | 5070.000000 | 5070.000000 | 5070.000000 | 5070.000000 | 5070.000000 | 5070.000000 | 5070.000000 | 5070.000000 | 5070.000000 | 5070.000000 |
| mean | 0.456805 | 37.986785 | 0.601183 | 89.423595 | 5.612839 | 4.114321 | 6.994371 | 0.381854 | 35.130769 | 3.301972 | 1.394477 |
| std | 0.498180 | 11.447095 | 0.764882 | 9.038394 | 2.257649 | 8.726001 | 13.651442 | 0.485889 | 8.919737 | 1.051700 | 0.510116 |
| min | 0.000000 | 0.000000 | 0.000000 | 30.000000 | -1.000000 | 0.000000 | 0.000000 | 0.000000 | 13.000000 | 0.000000 | 0.000000 |
| 25% | 0.000000 | 28.400000 | 0.000000 | 85.000000 | 4.314000 | 0.000000 | 0.000000 | 0.000000 | 27.000000 | 3.000000 | 1.000000 |
| 50% | 0.000000 | 36.550000 | 0.000000 | 89.000000 | 5.760000 | 0.000000 | 0.000000 | 0.000000 | 35.000000 | 4.000000 | 1.000000 |
| 75% | 1.000000 | 47.600000 | 1.000000 | 95.000000 | 7.193000 | 7.100000 | 4.120000 | 1.000000 | 42.000000 | 4.000000 | 2.000000 |
| max | 1.000000 | 65.900000 | 2.000000 | 126.000000 | 10.839000 | 108.960000 | 45.000000 | 1.000000 | 79.000000 | 4.000000 | 2.000000 |
4. 模型训练
4.1 LightGBM (0.96206)
首先尝试构建LightGBM模型。
Ref:
[1] Lightgbm原理、参数详解及python实例
[2] 深入理解LightGBM
以下为lightgbm采用5折交叉训练的代码:
#使用Lightgbm方法训练数据集,使用5折交叉验证的方法获得5个测试集预测结果
from sklearn.model_selection import KFold
from sklearn.model_selection import StratifiedKFold, GridSearchCV
def select_by_lgb(train_data,train_label,test_data,random_state=1234, n_splits=5,metric='auc',num_round=10000,early_stopping_rounds=100):
# kfold = KFold(n_splits=n_splits, shuffle=True, random_state=random_state)
kfold = StratifiedKFold(n_splits=n_splits, shuffle=True, random_state=random_state)
fold=0
result0 = []
for train_idx, val_idx in kfold.split(train_data, train_label):
random_state+=1
train_x = train_data.loc[train_idx]
train_y = train_label.loc[train_idx]
test_x = train_data.loc[val_idx]
test_y = train_label.loc[val_idx]
clf = lightgbm
train_matrix=clf.Dataset(train_x,label=train_y)
test_matrix=clf.Dataset(test_x,label=test_y)
params={
'boosting_type': 'gbdt',
'objective': 'binary',
'learning_rate': 0.1,
# 'max_depth': 7,
# 'num_leaves': 10,
'metric': metric,
'seed': random_state,
'silent': True,
'nthread':-1 }
model=clf.train(params,train_matrix,num_round,valid_sets=test_matrix,early_stopping_rounds=early_stopping_rounds)
pre_y=model.predict(test_data)
result0.append(pre_y)
fold+=1
pred_test = pd.DataFrame(result0).T
# 将5次预测结果求平均值
pred_test['average'] = pred_test.mean(axis=1)
#因为竞赛需要你提交最后的预测判断,而模型给出的预测结果是概率,因此我们认为概率>0.5的即该患者有糖尿病,概率<=0.5的没有糖尿病
pred_test['label'] = pred_test['average'].apply(lambda x:1 if x>0.5 else 0)
## 导出结果
result = pd.read_csv('提交示例.csv')
result['label']=pred_test['label']
return result
后面其他模型也需要进行k折交叉训练,这里定义一个k折交叉训练的函数,方便后续调用。
from sklearn.model_selection import KFold, StratifiedKFold
from sklearn.metrics import roc_auc_score, f1_score
def SKFold(train_data,train_label,test_data, model, random_state=1234, n_splits=5,metric='auc',num_round=10000,early_stopping_rounds=100):
# 采用分层K折交叉验证训练模型。
kfold = StratifiedKFold(n_splits=n_splits, shuffle=True, random_state=random_state)
fold = 1
pred_test = []
for train_idx, val_idx in kfold.split(train_data, train_label):
random_state+=1
train_x = train_data.loc[train_idx]
train_y = train_label.loc[train_idx]
val_x = train_data.loc[val_idx]
val_y = train_label.loc[val_idx]
eval_set = (val_x, val_y)
clf = model
model_trained = clf.fit(train_x, train_y)
# model_trained = clf.fit(train_x,train_y,early_stopping_rounds=early_stopping_rounds, verbose=False)
# model_trained = clf.fit(train_x, train_y, eval_set=eval_set, early_stopping_rounds=early_stopping_rounds)
pre_y = model_trained.predict(test_data)
pred_test.append(pre_y)
auc_train = roc_auc_score(train_y, model_trained.predict(train_x))
auc_val = roc_auc_score(val_y, model_trained.predict(val_x))
f_score_train = f1_score(train_y, model_trained.predict(train_x))
f_score_val = f1_score(val_y, model_trained.predict(val_x))
print('Fold: %d, AUC_train: %.4f, AUC_val: %.4f, F1-score_train: %.4f, F1-score_val: %.4f'%(fold,
auc_train, auc_val, f_score_train, f_score_val))
fold += 1
pred_test = pd.DataFrame(pred_test).T
# 将5次预测结果求平均值
pred_test['average'] = pred_test.mean(axis=1)
#因为竞赛需要你提交最后的预测判断,而模型给出的预测结果是概率,因此我们认为概率>0.5的即该患者有糖尿病,概率<=0.5的没有糖尿病
pred_test['label'] = pred_test['average'].apply(lambda x:1 if x>0.5 else 0)
## 导出结果
result=pd.read_csv('提交示例.csv')
result['label']=pred_test['label']
return result
由于比赛的测试集未公布,我们只能提交预测结果然后得到测试集上的分数,这里以表现较好的lightgbm模型作为baseline,若与lightgbm的预测结果相差较多则说明该模型表现不行。
def evaluate(result_LightGBM, result_others):
# 以lightGBM的结果为基准,评估其他模型的表现。
c = result_LightGBM['label'] - result_others['label']
count = 0
for i in c:
if i != 0:
count += 1
print('与LightGBM预测不同的样本数: ', count)
print(c[c!=0])
return count
先用select_by_lgb快速跑出一个baseline,然后用网格搜索得到最优参数,接着用最优参数组合在训练一遍模型,最后将结果提交。
random_state = 1234
result_LightGBM = select_by_lgb(X_train, Y_train, X_test) #baseline
result_LightGBM.to_csv('result_lightGBM.csv',index=False)
# 试试网格搜索最优参数
import lightgbm as lgb
params_test = {
'max_depth': range(4, 10, 1),
'num_leaves': range(10, 60, 10)
}
skf = StratifiedKFold(n_splits=5)
gsearch1 = GridSearchCV(estimator=lgb.LGBMClassifier(boosting_type='gbdt',objective='binary',metrics='auc',learning_rate=0.1, n_estimators=325,
max_depth=8, bagging_fraction = 0.8,feature_fraction = 0.8), param_grid=params_test,
scoring='roc_auc', cv=skf, n_jobs=-1)
gsearch1.fit(X_train, Y_train)
print(gsearch1.best_params_)
print(gsearch1.best_score_)
# 用最优参数再训练一遍
model_lgb = lgb.LGBMClassifier(boosting_type='gbdt',objective='binary',metrics='auc',
learning_rate=0.1, n_estimators=200, num_leaves=10, silent=True,
max_depth=7)
result_SKFold_lgb = SKFold(X_train, Y_train, X_test, model_lgb, n_splits=5)
result_SKFold_lgb.to_csv('result_SKFold_lgb.csv',index=False)
diff_lgb = evaluate(result_LightGBM, result_SKFold_lgb)
[LightGBM] [Warning] Unknown parameter: silent
[LightGBM] [Warning] Unknown parameter: silent
[LightGBM] [Info] Number of positive: 1549, number of negative: 2507
[LightGBM] [Warning] Auto-choosing row-wise multi-threading, the overhead of testing was 0.000386 seconds.
You can set `force_row_wise=true` to remove the overhead.
And if memory is not enough, you can set `force_col_wise=true`.
[LightGBM] [Info] Total Bins 1047
[LightGBM] [Info] Number of data points in the train set: 4056, number of used features: 10
[LightGBM] [Warning] Unknown parameter: silent
[LightGBM] [Info] [binary:BoostFromScore]: pavg=0.381903 -> initscore=-0.481477
[LightGBM] [Info] Start training from score -0.481477
[1] valid_0's auc: 0.979322
Training until validation scores don't improve for 100 rounds
[2] valid_0's auc: 0.980354
[3] valid_0's auc: 0.982351
[4] valid_0's auc: 0.981993
Early stopping, best iteration is:
[40] valid_0's auc: 0.989851
[LightGBM] [Warning] feature_fraction is set=0.8, colsample_bytree=1.0 will be ignored. Current value: feature_fraction=0.8
[LightGBM] [Warning] bagging_fraction is set=0.8, subsample=1.0 will be ignored. Current value: bagging_fraction=0.8
{'max_depth': 7, 'num_leaves': 10}
0.9902553653233458
Fold: 1, AUC_train: 0.9917, AUC_val: 0.9461, F1-score_train: 0.9909, F1-score_val: 0.9358
[LightGBM] [Warning] feature_fraction is set=0.8, colsample_bytree=1.0 will be ignored. Current value: feature_fraction=0.8
[LightGBM] [Warning] bagging_fraction is set=0.8, subsample=1.0 will be ignored. Current value: bagging_fraction=0.8
Fold: 2, AUC_train: 0.9852, AUC_val: 0.9487, F1-score_train: 0.9837, F1-score_val: 0.9386
[LightGBM] [Warning] feature_fraction is set=0.8, colsample_bytree=1.0 will be ignored. Current value: feature_fraction=0.8
[LightGBM] [Warning] bagging_fraction is set=0.8, subsample=1.0 will be ignored. Current value: bagging_fraction=0.8
Fold: 3, AUC_train: 0.9863, AUC_val: 0.9457, F1-score_train: 0.9853, F1-score_val: 0.9328
[LightGBM] [Warning] feature_fraction is set=0.8, colsample_bytree=1.0 will be ignored. Current value: feature_fraction=0.8
[LightGBM] [Warning] bagging_fraction is set=0.8, subsample=1.0 will be ignored. Current value: bagging_fraction=0.8
Fold: 4, AUC_train: 0.9876, AUC_val: 0.9607, F1-score_train: 0.9860, F1-score_val: 0.9490
[LightGBM] [Warning] feature_fraction is set=0.8, colsample_bytree=1.0 will be ignored. Current value: feature_fraction=0.8
[LightGBM] [Warning] bagging_fraction is set=0.8, subsample=1.0 will be ignored. Current value: bagging_fraction=0.8
Fold: 5, AUC_train: 0.9858, AUC_val: 0.9505, F1-score_train: 0.9841, F1-score_val: 0.9403
与LightGBM预测不同的样本数: 7
24 1
35 -1
43 1
76 1
434 1
442 1
501 -1
Name: label, dtype: int64
4.2 随机森林(0.96324)
Ref:
[1] Permutation Importance vs Random Forest Feature Importance (MDI)
from sklearn.ensemble import RandomForestClassifier
forest = RandomForestClassifier(max_depth=5, random_state=1234)
forest.fit(X_train, Y_train)
pred_forest = forest.predict(X_test)
result=pd.read_csv('提交示例.csv')
result['label']=pred_forest
result.to_csv('result_RandomForest.csv',index=False)
feature_importance_forest = pd.Series(forest.feature_importances_,
index=X_train.columns).sort_values(ascending=True)
plt.figure(figsize=(10, 7), dpi=80)
ax = feature_importance_forest.plot.barh()
ax.set_title("Random Forest Feature Importances (MDI)")
# ax.figure.tight_layout()
## 网格搜索最优参数组合
params_test = {
'max_depth': range(3, 20, 2),
'n_estimators': range(100, 600, 100),
'min_samples_leaf': [2, 4, 6]
}
skf = StratifiedKFold(n_splits=5)
gsearch2 = GridSearchCV(estimator=RandomForestClassifier(n_estimators=200, max_depth=5, random_state=random_state),
param_grid=params_test, scoring='roc_auc', cv=skf, n_jobs=-1)
gsearch2.fit(X_train, Y_train)
print(gsearch2.best_params_)
print(gsearch2.best_score_)
{'max_depth': 13, 'min_samples_leaf': 2, 'n_estimators': 400}
0.9927105570462718
model_forest = RandomForestClassifier(n_estimators=400, max_depth=13, random_state=random_state)
result_SKFold_forest = SKFold(X_train, Y_train, X_test, model_forest)
result_SKFold_forest.to_csv('result_skfold_RandomForest.csv',index=False)
diff_skold_forest = evaluate(result_LightGBM, result_SKFold_forest)
Fold: 1, AUC_train: 0.9935, AUC_val: 0.9516, F1-score_train: 0.9935, F1-score_val: 0.9424
Fold: 2, AUC_train: 0.9910, AUC_val: 0.9564, F1-score_train: 0.9909, F1-score_val: 0.9468
Fold: 3, AUC_train: 0.9919, AUC_val: 0.9490, F1-score_train: 0.9919, F1-score_val: 0.9396
Fold: 4, AUC_train: 0.9919, AUC_val: 0.9647, F1-score_train: 0.9919, F1-score_val: 0.9551
Fold: 5, AUC_train: 0.9913, AUC_val: 0.9591, F1-score_train: 0.9912, F1-score_val: 0.9497
与LightGBM预测不同的样本数: 11
8 1
35 -1
47 -1
52 -1
60 1
64 1
76 1
78 1
85 1
618 -1
796 -1
Name: label, dtype: int64
4.3 XGBoost (0.95981)
Ref:
[1] XGBoost:在Python中使用XGBoost
[2] Python机器学习笔记:XgBoost算法
[3] python包xgboost安装和简单使用
[4] 深入理解XGBoost,优缺点分析,原理推导及工程实现
[5] XGBoost的原理、公式推导、Python实现和应用
[6] XGBoost官方文档
import xgboost as xgb
from xgboost import plot_importance
from sklearn.metrics import roc_auc_score
from sklearn.model_selection import StratifiedKFold
import warnings
warnings.filterwarnings('ignore')
# 分层k折交叉检验
skf = StratifiedKFold(n_splits=5)
result_xgb = []
fold = 1
for train_idx, val_idx in skf.split(X_train, Y_train):
train_x = X_train.loc[train_idx]
train_y = Y_train.loc[train_idx]
val_x = X_train.loc[val_idx]
val_y = Y_train.loc[val_idx]
d_train = xgb.DMatrix(train_x, train_y)
d_val = xgb.DMatrix(val_x, val_y)
d_test = xgb.DMatrix(X_test)
params = {
'max_depth':5,
'min_child_weight':1,
'num_class':2,
'eta': 0.1, #学习率
'gamma': 0.1, #后剪枝参数,取值在[0, 1],越大越保守
'seed': 1234,
'alpha': 1, #L1正则项的惩罚系数
'eval_metric': 'auc'
}
num_round = 500
# # 方式一:采用sklearn接口,采用fit 和 predict
# model_xgb = xgb.XGBClassifier()
# model_xgb.fit(train_x, train_y, verbose=False)
# pred_train = model_xgb.predict(train_x)
# pred_val = model_xgb.predict(val_x)
# pred_xgb = model_xgb.predict(X_test)
# 方式二:采用xgboost原生接口,采用train和predict,方便调参
model_xgb = xgb.train(params, d_train, num_round)
pred_train = model_xgb.predict(d_train)
pred_val = model_xgb.predict(d_val)
pred_xgb = model_xgb.predict(d_test)
auc_train = roc_auc_score(train_y, pred_train)
auc_val = roc_auc_score(val_y, pred_val)
f_score_train = f1_score(train_y, pred_train)
f_score_val = f1_score(val_y, pred_val)
print('Fold: %d, AUC_train: %.4f, AUC_val: %.4f, F1-score_train: %.4f, F1-score_val: %.4f'%(fold,
auc_train, auc_val, f_score_train, f_score_val))
result_xgb.append(pred_xgb)
fold += 1
result_xgb = pd.DataFrame(result_xgb).T
print('result_xgb.shape = ', result_xgb.shape)
# 将5次预测结果求平均值
result_xgb['average'] = result_xgb.mean(axis=1)
# 最终预测结果
result_xgb['label'] = result_xgb['average'].apply(lambda x:1 if x>0.5 else 0)
# 特征重要性
plot_importance(model_xgb)
plt.show()
# 导出结果
result = pd.read_csv('提交示例.csv')
result['label'] = result_xgb['label']
result.to_csv('result_XGBoost_StratifiedKFold.csv',index=False)
diff_xgb = evaluate(result_LightGBM, result_xgb)
Fold: 1, AUC_train: 0.9935, AUC_val: 0.9463, F1-score_train: 0.9929, F1-score_val: 0.9349
Fold: 2, AUC_train: 0.9952, AUC_val: 0.9556, F1-score_train: 0.9948, F1-score_val: 0.9456
Fold: 3, AUC_train: 0.9952, AUC_val: 0.9518, F1-score_train: 0.9948, F1-score_val: 0.9415
Fold: 4, AUC_train: 0.9906, AUC_val: 0.9524, F1-score_train: 0.9893, F1-score_val: 0.9407
Fold: 5, AUC_train: 0.9924, AUC_val: 0.9573, F1-score_train: 0.9919, F1-score_val: 0.9482
result_xgb.shape = (1000, 5)
与LightGBM预测不同的样本数: 6
21 -1
24 1
28 -1
35 -1
43 1
74 -1
Name: label, dtype: int64
4.4 CatBoost(0.95854)
CatbBoost 是GBDT算法框架的一种改进实现,其主要创新点有:
- 支持类别性变量。嵌入了自动将类别型特征处理为数值型特征的创新算法。
- 使用了组合类别特征,丰富特征维度。
- 采用排序提升的方法对抗训练集中的噪声点,从而避免梯度估计的偏差,进而解决预测偏移的问题。
- 采用了完全对称树作为基模型。
Ref:
[1] 深入理解CatBoost
[2] Catboost 一个超级简单实用的boost算法
import catboost as cb
from sklearn.metrics import roc_auc_score
from sklearn.model_selection import StratifiedKFold
# 分层k折交叉检验
skf = StratifiedKFold(n_splits=5)
categorical_features_index = np.where(X_train.dtypes != float)[0]
print(X_train.columns[categorical_features_index])
result_cat = []
fold = 1
for train_idx, val_idx in skf.split(X_train, Y_train):
train_x = X_train.loc[train_idx]
train_y = Y_train.loc[train_idx]
val_x = X_train.loc[val_idx]
val_y = Y_train.loc[val_idx]
model_catboost = cb.CatBoostClassifier(eval_metric='AUC', cat_features=categorical_features_index,
depth=6, n_estimators=400, learning_rate=0.5, verbose=False)
model_catboost.fit(train_x, train_y, eval_set=(val_x, val_y), plot=False)
pred_train = model_catboost.predict(train_x)
pred_val = model_catboost.predict(val_x)
auc_train = roc_auc_score(train_y, pred_train)
auc_val = roc_auc_score(val_y, pred_val)
f_score_train = f1_score(train_y, pred_train)
f_score_val = f1_score(val_y, pred_val)
print('Fold: %d, AUC_train: %.4f, AUC_val: %.4f, F1-score_train: %.4f, F1-score_val: %.4f'%(fold,
auc_train, auc_val, f_score_train, f_score_val))
pred_catboost = model_catboost.predict(X_test)
result_cat.append(pred_catboost)
fold += 1
result_cat = pd.DataFrame(result_cat).T
print('result_cat.shape = ', result_cat.shape)
# 将5次预测结果求平均值
result_cat['average'] = result_cat.mean(axis=1)
# 最终预测结果
result_cat['label'] = result_cat['average'].apply(lambda x:1 if x>0.5 else 0)
# 导出结果
result = pd.read_csv('提交示例.csv')
result['label'] = result_cat['label']
result.to_csv('result_CatBoost_StratifiedKFold.csv',index=False)
diff_catboost = evaluate(result_LightGBM, result_cat)
feature_importance_catboost = model_catboost.feature_importances_
plt.figure(figsize=(10,8), dpi=80)
plt.barh(col_names, feature_importance_catboost)
plt.show()
Index(['性别', '糖尿病家族史', 'BMI', 'DBP'], dtype='object')
Fold: 1, AUC_train: 0.9902, AUC_val: 0.9419, F1-score_train: 0.9887, F1-score_val: 0.9305
Fold: 2, AUC_train: 0.9775, AUC_val: 0.9608, F1-score_train: 0.9722, F1-score_val: 0.9510
Fold: 3, AUC_train: 0.9988, AUC_val: 0.9543, F1-score_train: 0.9984, F1-score_val: 0.9442
Fold: 4, AUC_train: 0.9590, AUC_val: 0.9448, F1-score_train: 0.9498, F1-score_val: 0.9298
Fold: 5, AUC_train: 0.9651, AUC_val: 0.9484, F1-score_train: 0.9578, F1-score_val: 0.9377
result_cat.shape = (1000, 5)
与LightGBM预测不同的样本数: 16
8 1
33 -1
35 -1
47 -1
52 -1
60 1
64 1
74 -1
76 1
83 -1
85 1
89 -1
166 -1
501 -1
618 -1
796 -1
Name: label, dtype: int64
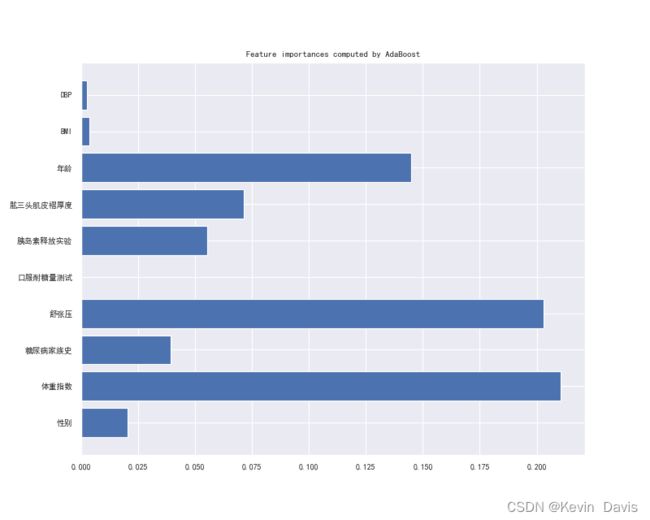
4.5 AdaBoost(0.96098)
from sklearn.ensemble import AdaBoostClassifier
from sklearn.tree import DecisionTreeClassifier
random_state = 1234
model_tree = DecisionTreeClassifier(max_depth=5, random_state=random_state)
model_adaboost = AdaBoostClassifier(base_estimator=model_tree, n_estimators=200,
random_state=random_state)
result_adaboost = SKFold(X_train, Y_train, X_test, model_adaboost)
result_adaboost.to_csv('result_AdaBoost.csv',index=False)
# 评估
diff_skold_adaboost = evaluate(result_LightGBM, result_adaboost)
# 特征重要性
feature_importance_adaboost = model_adaboost.feature_importances_
plt.figure(figsize=(10,8), dpi=80)
plt.rc('font', size = 18)
plt.barh(col_names, feature_importance_adaboost)
plt.title('Feature importances computed by AdaBoost')
plt.show()
Fold: 1, AUC_train: 1.0000, AUC_val: 0.9458, F1-score_train: 1.0000, F1-score_val: 0.9347
Fold: 2, AUC_train: 1.0000, AUC_val: 0.9445, F1-score_train: 1.0000, F1-score_val: 0.9333
Fold: 3, AUC_train: 1.0000, AUC_val: 0.9380, F1-score_train: 1.0000, F1-score_val: 0.9263
Fold: 4, AUC_train: 1.0000, AUC_val: 0.9488, F1-score_train: 1.0000, F1-score_val: 0.9357
Fold: 5, AUC_train: 1.0000, AUC_val: 0.9536, F1-score_train: 1.0000, F1-score_val: 0.9432
与LightGBM预测不同的样本数: 12
35 -1
47 -1
50 -1
52 -1
60 1
64 1
76 1
85 1
92 -1
94 1
495 1
796 -1
Name: label, dtype: int64
4.6 集成模型(0.95971)
挑几个表现较好的模型进行集成。
%%time
skf = StratifiedKFold(n_splits=5)
categorical_features_index = np.where(X_train.dtypes != float)[0]
print('类别型特征: ', X_train.columns[categorical_features_index])
cat_features = list(map(lambda x:int(x), categorical_features_index))
random_state = 1234
fold = 1
for train_idx, val_idx in skf.split(X_train, Y_train):
train_x = X_train.loc[train_idx]
train_y = Y_train.loc[train_idx]
val_x = X_train.loc[val_idx]
val_y = Y_train.loc[val_idx]
d_train = xgb.DMatrix(train_x, train_y)
d_val = xgb.DMatrix(val_x, val_y)
d_test = xgb.DMatrix(X_test)
params_xgb = {
'max_depth':5,
'min_child_weight':1,
'num_class':2,
'eta': 0.1, #学习率
'gamma': 0.1, #后剪枝参数,取值在[0, 1],越大越保守
'seed': 1234,
'alpha': 1, #L1正则项的惩罚系数
'eval_metric': 'auc'
}
num_round = 500
early_stopping_rounds = 100
model_lightGBM = lgb.LGBMClassifier(boosting_type='gbdt',objective='binary',metrics='auc',
learning_rate=0.1, n_estimators=200, num_leaves=10, silent=True,
max_depth=7)
model_lightGBM.fit(X_train, Y_train)
model_forest = RandomForestClassifier(max_depth=13, n_estimators=400, random_state=1234)
model_forest.fit(X_train, Y_train)
model_tree = DecisionTreeClassifier(max_depth=5, random_state=random_state)
model_adaboost = AdaBoostClassifier(base_estimator=model_tree, n_estimators=200,
random_state=random_state)
model_adaboost.fit(X_train, Y_train)
model_xgb = xgb.train(params_xgb, d_train, num_round)
model_catboost = cb.CatBoostClassifier(eval_metric='AUC', cat_features=categorical_features_index,
depth=6, iterations=400, learning_rate=0.5, verbose=False)
model_catboost.fit(train_x, train_y, eval_set=(val_x, val_y), plot=False)
print('Fold: %d finished training. '%fold)
fold += 1
pred_lightGBM = model_lightGBM.predict(test_data)
# pred_lightGBM = list(map(lambda x: 1 if x>0.5 else 0, pred_lightGBM)) #调用lightGBM原生接口时使用
pred_forest = forest.predict(X_test)
pred_adaboost = model_adaboost.predict(X_test)
pred_xgb = model_xgb.predict(d_test)
pred_catboost = model_catboost.predict(X_test)
pred_all = pd.DataFrame({'lightGBM': pred_lightGBM,
'RandomForest': pred_forest,
'AdaBoost': pred_adaboost,
'XGBoost': pred_xgb,
'CatBoost': pred_catboost})
pred_all['Average'] = pred_all.mean(axis=1)
# 最终预测结果
pred_all['label'] = pred_all['Average'].apply(lambda x:1 if x>0.5 else 0)
# 导出结果
result = pd.read_csv('提交示例.csv')
result['label'] = pred_all['label']
result.to_csv('result_Ensemble.csv',index=False)
diff_ensemble = evaluate(result_LightGBM, result)
类别型特征: Index(['性别', '糖尿病家族史', 'BMI', 'DBP'], dtype='object')
Custom logger is already specified. Specify more than one logger at same time is not thread safe.
Fold: 1 finished training.
Fold: 2 finished training.
Fold: 3 finished training.
Fold: 4 finished training.
Fold: 5 finished training.
与LightGBM预测不同的样本数: 11
24 1
33 -1
35 -1
43 1
52 -1
60 1
64 1
76 1
78 1
85 1
796 -1
Name: label, dtype: int64
Wall time: 1min 31s
4.7 Stacking(0.96577)
Stacking的思想为在初始数据集上训练若干个基学习器,并将这几个基学习器的预测结果作为新的训练集,来训练一个新的学习器,并将其预测结果作为最终输出。
Stacking本质是一种层级结构,第一层有n个基学习器,每个基学习器进行k折交叉训练,把每一折的验证集(validation set)的预测结果输出并拼接在一起,把这n个模型的训练集预测结果作为新的训练集,将这n个模型的测试集预测结果拼接在一起作为新的测试集。
Ref:
[1] stacking模型融合
[2] Kaggle上分技巧——单模K折交叉验证训练+多模型融合
model_tree = DecisionTreeClassifier(max_depth=5, random_state=random_state)
clfs = [lgb.LGBMClassifier(boosting_type='gbdt',objective='binary',metrics='auc',
learning_rate=0.1, n_estimators=200, num_leaves=10, silent=True,
max_depth=7),
RandomForestClassifier(max_depth=13, n_estimators=400, random_state=1234),
AdaBoostClassifier(base_estimator=model_tree, n_estimators=200,
random_state=random_state),
xgb.XGBClassifier(),
cb.CatBoostClassifier(eval_metric='AUC', cat_features=categorical_features_index,
depth=6, iterations=400, learning_rate=0.5, verbose=False)]
data_train = np.zeros((X_train.shape[0], len(clfs)))
data_test = np.zeros((X_test.shape[0], len(clfs)))
# 5折stacking
n_splits = 5
skf = StratifiedKFold(n_splits)
# 第一层,训练各个个体学习器
for i, clf in enumerate(clfs):
# 依次训练各个模型
d_test = np.zeros((X_test.shape[0], n_splits)) #存放个体学习器在测试集上的预测输出
for fold, (train_idx, val_idx) in enumerate(skf.split(X_train, Y_train)):
#5折交叉训练,第j折拿来预测并作为第二层模型的训练集,剩余部分拿来训练模型。
train_x = X_train.loc[train_idx]
train_y = Y_train.loc[train_idx]
val_x = X_train.loc[val_idx]
val_y = Y_train.loc[val_idx]
clf.fit(train_x, train_y)
pred_train = clf.predict(train_x)
pred_val = clf.predict(val_x)
data_train[val_idx, i] = pred_val
d_test[:, fold] = clf.predict(X_test)
auc_train = roc_auc_score(train_y, pred_train)
auc_val = roc_auc_score(val_y, pred_val)
f_score_train = f1_score(train_y, pred_train)
f_score_val = f1_score(val_y, pred_val)
print('Classifier:%d, Fold: %d, AUC_train: %.4f, AUC_val: %.4f, F1-score_train: %.4f, F1-score_val: %.4f'%(i+1,
fold+1, auc_train, auc_val, f_score_train, f_score_val))
#对于测试集,直接用这交叉验证训练的每个模型的预测值均值作为新的特征
data_test[:, i] = d_test.mean(axis=1)
data_train = pd.DataFrame(data_train)
data_test = pd.DataFrame(data_test)
# 第二层改用高级点的模型,并进行5折交叉训练
# model_forest = RandomForestClassifier(max_depth=5, random_state=1234)
model_2 = lgb.LGBMClassifier(boosting_type='gbdt',objective='binary',metrics='auc',
learning_rate=0.3, n_estimators=200, num_leaves=10, silent=True,
max_depth=7)
result_stack = SKFold(data_train, Y_train, data_test, model_2)
result_stack.to_csv('result_stack.csv', index=False)
diff_stack = evaluate(result_LightGBM, result_stack)
Classifier:1, Fold: 1, AUC_train: 0.9894, AUC_val: 0.9465, F1-score_train: 0.9880, F1-score_val: 0.9340
Classifier:1, Fold: 2, AUC_train: 0.9883, AUC_val: 0.9554, F1-score_train: 0.9870, F1-score_val: 0.9465
Classifier:1, Fold: 3, AUC_train: 0.9886, AUC_val: 0.9559, F1-score_train: 0.9873, F1-score_val: 0.9467
Classifier:1, Fold: 4, AUC_train: 0.9897, AUC_val: 0.9417, F1-score_train: 0.9883, F1-score_val: 0.9268
Classifier:1, Fold: 5, AUC_train: 0.9887, AUC_val: 0.9542, F1-score_train: 0.9879, F1-score_val: 0.9453
Classifier:2, Fold: 1, AUC_train: 0.9926, AUC_val: 0.9489, F1-score_train: 0.9925, F1-score_val: 0.9377
Classifier:2, Fold: 2, AUC_train: 0.9929, AUC_val: 0.9577, F1-score_train: 0.9928, F1-score_val: 0.9482
Classifier:2, Fold: 3, AUC_train: 0.9923, AUC_val: 0.9562, F1-score_train: 0.9922, F1-score_val: 0.9478
Classifier:2, Fold: 4, AUC_train: 0.9929, AUC_val: 0.9569, F1-score_train: 0.9928, F1-score_val: 0.9470
Classifier:2, Fold: 5, AUC_train: 0.9903, AUC_val: 0.9529, F1-score_train: 0.9902, F1-score_val: 0.9439
Classifier:3, Fold: 1, AUC_train: 1.0000, AUC_val: 0.9377, F1-score_train: 1.0000, F1-score_val: 0.9253
Classifier:3, Fold: 2, AUC_train: 1.0000, AUC_val: 0.9478, F1-score_train: 1.0000, F1-score_val: 0.9354
Classifier:3, Fold: 3, AUC_train: 1.0000, AUC_val: 0.9554, F1-score_train: 1.0000, F1-score_val: 0.9465
Classifier:3, Fold: 4, AUC_train: 1.0000, AUC_val: 0.9395, F1-score_train: 1.0000, F1-score_val: 0.9269
Classifier:3, Fold: 5, AUC_train: 1.0000, AUC_val: 0.9495, F1-score_train: 1.0000, F1-score_val: 0.9399
[00:31:58] WARNING: C:/Users/Administrator/workspace/xgboost-win64_release_1.5.0/src/learner.cc:1115: Starting in XGBoost 1.3.0, the default evaluation metric used with the objective 'binary:logistic' was changed from 'error' to 'logloss'. Explicitly set eval_metric if you'd like to restore the old behavior.
Classifier:4, Fold: 1, AUC_train: 0.9997, AUC_val: 0.9452, F1-score_train: 0.9997, F1-score_val: 0.9326
[00:31:59] WARNING: C:/Users/Administrator/workspace/xgboost-win64_release_1.5.0/src/learner.cc:1115: Starting in XGBoost 1.3.0, the default evaluation metric used with the objective 'binary:logistic' was changed from 'error' to 'logloss'. Explicitly set eval_metric if you'd like to restore the old behavior.
Classifier:4, Fold: 2, AUC_train: 0.9997, AUC_val: 0.9530, F1-score_train: 0.9997, F1-score_val: 0.9429
[00:31:59] WARNING: C:/Users/Administrator/workspace/xgboost-win64_release_1.5.0/src/learner.cc:1115: Starting in XGBoost 1.3.0, the default evaluation metric used with the objective 'binary:logistic' was changed from 'error' to 'logloss'. Explicitly set eval_metric if you'd like to restore the old behavior.
Classifier:4, Fold: 3, AUC_train: 1.0000, AUC_val: 0.9538, F1-score_train: 1.0000, F1-score_val: 0.9441
[00:31:59] WARNING: C:/Users/Administrator/workspace/xgboost-win64_release_1.5.0/src/learner.cc:1115: Starting in XGBoost 1.3.0, the default evaluation metric used with the objective 'binary:logistic' was changed from 'error' to 'logloss'. Explicitly set eval_metric if you'd like to restore the old behavior.
Classifier:4, Fold: 4, AUC_train: 0.9994, AUC_val: 0.9480, F1-score_train: 0.9994, F1-score_val: 0.9345
[00:32:00] WARNING: C:/Users/Administrator/workspace/xgboost-win64_release_1.5.0/src/learner.cc:1115: Starting in XGBoost 1.3.0, the default evaluation metric used with the objective 'binary:logistic' was changed from 'error' to 'logloss'. Explicitly set eval_metric if you'd like to restore the old behavior.
Classifier:4, Fold: 5, AUC_train: 0.9994, AUC_val: 0.9537, F1-score_train: 0.9994, F1-score_val: 0.9452
Classifier:5, Fold: 1, AUC_train: 0.9980, AUC_val: 0.9411, F1-score_train: 0.9977, F1-score_val: 0.9293
Classifier:5, Fold: 2, AUC_train: 0.9992, AUC_val: 0.9587, F1-score_train: 0.9990, F1-score_val: 0.9485
Classifier:5, Fold: 3, AUC_train: 0.9990, AUC_val: 0.9556, F1-score_train: 0.9987, F1-score_val: 0.9456
Classifier:5, Fold: 4, AUC_train: 0.9997, AUC_val: 0.9464, F1-score_train: 0.9997, F1-score_val: 0.9321
Classifier:5, Fold: 5, AUC_train: 0.9987, AUC_val: 0.9447, F1-score_train: 0.9987, F1-score_val: 0.9326
Fold: 1, AUC_train: 0.9582, AUC_val: 0.9455, F1-score_train: 0.9491, F1-score_val: 0.9337
Fold: 2, AUC_train: 0.9538, AUC_val: 0.9495, F1-score_train: 0.9449, F1-score_val: 0.9398
Fold: 3, AUC_train: 0.9569, AUC_val: 0.9471, F1-score_train: 0.9477, F1-score_val: 0.9361
Fold: 4, AUC_train: 0.9546, AUC_val: 0.9556, F1-score_train: 0.9451, F1-score_val: 0.9456
Fold: 5, AUC_train: 0.9544, AUC_val: 0.9518, F1-score_train: 0.9463, F1-score_val: 0.9416
与LightGBM预测不同的样本数: 14
0 -1
8 1
23 -1
28 -1
33 -1
35 -1
47 -1
52 -1
60 1
64 1
74 -1
89 -1
796 -1
851 -1
Name: label, dtype: int64
4.8 归一化数据,pytorch神经网络
先归一化数据,统一量纲。从本节开始,使用基于距离的模型,不再使用树模型。
注意pytorch做二元分类有以下几种实现方式:
- Linear 输出维度为1 + sigmoid + BCELoss。
- Linear 输出维度为1 + BCEWithLogitsLoss。不需要加sigmoid或softmax函数,BCEWithLogitsLoss自带sigmoid作为激活函数。
- Linear 输出维度为2 + 交叉熵(CrossEntropyLoss)。输出tensor的维度0对应第一个label(即0),维度1对应第二个label(即1)。注意使用交叉熵时,真实标签不能是onehot格式,必须为1维tensor,预测标签必须大于或等于2维,预测标签的每一个维度对应一个标签。
pytorch 中使用神经网络进行多分类时,网络的输出 prediction 是 one hot 格式,但计算 交叉熵损失函数时,loss = criterion(prediction, target) 的输入 target 不能是 one hot 格式,直接用数字来表示就行(4 表示 one hot 中的 0 0 0 1)。
所以,自己构建数据集,返回的 target 不需要是 one hot 格式。
Ref:
[1] Pytorch学习笔记(5)——交叉熵报错RuntimeError: 1D target tensor expected, multi-target not supported
[2] PyTorch二分类时BCELoss,CrossEntropyLoss,Sigmoid等的选择和使用
[3] Pytorch实现二分类器
[4] RuntimeError: multi-target not supported at
from sklearn.preprocessing import MinMaxScaler
# 归一化
scaler = MinMaxScaler()
X_train2 = scaler.fit_transform(X_train)
X_test2 = scaler.fit_transform(X_test)
Y_train2 = Y_train.to_numpy()
print('X_train.shape = ', X_train.shape)
print('X_train2.shape = ', X_train2.shape)
print('Y_train2.shape = ', Y_train2.shape)
X_train.shape = (5070, 10)
X_train2.shape = (5070, 10)
Y_train2.shape = (5070,)
def Convert(x):
# Conver the numeric values into categorical values.
y = np.zeros((x.shape[0],))
for i in range(len(x)):
if x[i, 0] > x[i, 1]:
y[i] = 0
else:
y[i] = 1
return y
import torch
import torch.nn as nn
import torch.nn.functional as F
class NET(nn.Module):
def __init__(self, input_dim:int, hidden:int, out_dim:int, activation='relu', dropout=0.2):
super(NET, self).__init__()
self.input_dim = input_dim
self.hidden = hidden
self.out_dim = out_dim
self.activation = activation
self.Dropout = dropout
# 激活函数选择
if self.activation == 'relu':
mid_act = torch.nn.ReLU()
elif self.activation == 'tanh':
mid_act = torch.nn.Tanh()
elif self.activation == 'sigmoid':
mid_act = torch.nn.Sigmoid()
elif self.activation == 'LeakyReLU':
mid_act = torch.nn.LeakyReLU()
elif self.activation == 'ELU':
mid_act = torch.nn.ELU()
elif self.activation == 'GELU':
mid_act = torch.nn.GELU()
self.model = nn.Sequential(
nn.Linear(self.input_dim, self.hidden),
mid_act,
nn.Dropout(self.Dropout),
nn.Linear(self.hidden, self.hidden),
mid_act,
nn.Dropout(self.Dropout),
nn.Linear(hidden, self.out_dim)
)
def forward(self, x):
out = self.model(x)
return out
def predict(self, x):
# x = torch.tensor(x.to_numpy()) #针对datafram
# x = x.to(torch.float32)
x = torch.tensor(x).to(torch.float32) #针对ndarray
x = F.softmax(self.model(x))
ans = []
for t in x:
if t[0] > t[1]:
ans.append(0)
else:
ans.append(1)
return np.array(ans)
import time
from torch.utils.data import DataLoader, TensorDataset
class NN_classifier():
def __init__(self, model, crit, l_rate, batch_size, max_epochs, n_splits=5, verbose=True):
super(NN_classifier, self).__init__()
self.model = model # Neural network model, should be a nn.Module()
self.l_rate = l_rate
self.batch_size = batch_size
self.max_epochs = max_epochs
self.verbose = verbose
self.n_splits = n_splits # the value of k in k-fold validation
self.crit = crit # loss function
self.device = 'cpu'
def fit(self, X_train, Y_train, X_test):
skf = StratifiedKFold(n_splits=self.n_splits)
fold = 1
pred_test = []
for train_idx, val_idx in skf.split(X_train, Y_train):
train_x = X_train[train_idx, :]
train_y = Y_train[train_idx]
val_x = X_train[val_idx, :]
val_y = Y_train[val_idx]
train_data = TensorDataset(train_x, train_y)
train_dataloader = DataLoader(dataset=train_data, batch_size=self.batch_size, shuffle=True)
valid_data = TensorDataset(val_x, val_y)
validation_dataloader = DataLoader(dataset=valid_data, batch_size=self.batch_size, shuffle=False)
model = self.model
optimizer = torch.optim.Adam(model.parameters(), lr=self.l_rate)
scheduler = torch.optim.lr_scheduler.StepLR(optimizer, step_size=50, gamma=0.9) #动态学习率调整
for epoch in range(self.max_epochs):
start_time = time.time()
loss_all = []
#------------- Training -----------------
model.train()
for data in train_dataloader:
x, y = data
x = x.to(self.device)
y = y.to(self.device)
optimizer.zero_grad()
out = model(x)
loss = self.crit(out, y.long())
loss.requires_grad_(True)
loss.backward()
optimizer.step()
loss_all.append(loss.item())
scheduler.step()
end_time = time.time()
cost_time = end_time - start_time
train_loss = np.mean(np.array(loss_all))
#------------- Validation -----------------
model.eval()
loss_all = []
with torch.no_grad():
for data in validation_dataloader:
x, y = data
x = x.to(self.device)
y = y.to(self.device)
output = model(x)
loss = self.crit(output, y.long())
loss_all.append(loss.item())
validation_loss = np.mean(np.array(loss_all))
if self.verbose and (epoch+1) % 100 ==0:
print('Fold:{:d}, Epoch:{:d}, train_loss: {:.4f}, validation_loss: {:.4f}, cost_time: {:.2f}s'
.format(fold, epoch+1, train_loss, validation_loss, cost_time))
#------------- Prediction -----------------
pred = Convert(model(X_test).detach().numpy())
pred_test.append(pred)
pred_train = Convert(model(train_x).detach().numpy())
pred_val = Convert(model(val_x).detach().numpy())
auc_train = roc_auc_score(train_y, pred_train)
auc_val = roc_auc_score(val_y, pred_val)
f_score_train = f1_score(train_y, pred_train)
f_score_val = f1_score(val_y, pred_val)
print('Fold: %d, AUC_train: %.4f, AUC_val: %.4f, F1-score_train: %.4f, F1-score_val: %.4f'%(fold,
auc_train, auc_val, f_score_train, f_score_val))
fold += 1
pred_test = pd.DataFrame(pred_test).T
print('pred_test.shape = ', pred_test.shape)
# 将5次预测结果求平均值
pred_test['average'] = pred_test.mean(axis=1)
#因为竞赛需要你提交最后的预测判断,而模型给出的预测结果是概率,因此我们认为概率>0.5的即该患者有糖尿病,概率<=0.5的没有糖尿病
pred_test['label'] = pred_test['average'].apply(lambda x:1 if x>0.5 else 0)
## 导出结果
result=pd.read_csv('提交示例.csv')
result['label']=pred_test['label']
return result
# k折交叉验证不断训练同一个模型,集成不同fold(即不同时刻)的模型的预测结果
hidden = 64
activation = 'relu'
# activation = 'tanh'
# crit = nn.MSELoss()
crit = nn.CrossEntropyLoss()
batch_size = 512*2
max_epochs = 500
l_rate = 1e-3
dropout = 0.1
n_splits = 5
# Convert to tensor
X_train_tensor = torch.from_numpy(X_train2).to(torch.float32)
X_test_tensor = torch.from_numpy(X_test2).to(torch.float32)
Y_train_tensor = torch.from_numpy(Y_train2).to(torch.float32)
model_NN = NET(X_train2.shape[1], hidden, out_dim=2, activation=activation)
classifier_NN = NN_classifier(model_NN, crit=crit, batch_size=batch_size, l_rate=l_rate,
max_epochs=max_epochs, n_splits=n_splits)
result_SKFold_NN = NN_classifier.fit(classifier_NN,X_train_tensor, Y_train_tensor, X_test_tensor,)
c = result_LightGBM['label'] - result_SKFold_NN['label']
count = 0
for i in c:
if i != 0:
count += 1
print('与LightGBM预测不同的样本数: ', count)
print(c[c!=0])
Fold:1, Epoch:100, train_loss: 0.3503, validation_loss: 0.3196, cost_time: 0.04s
Fold:1, Epoch:200, train_loss: 0.2785, validation_loss: 0.2505, cost_time: 0.04s
Fold:1, Epoch:300, train_loss: 0.2332, validation_loss: 0.2257, cost_time: 0.04s
Fold:1, Epoch:400, train_loss: 0.2098, validation_loss: 0.2130, cost_time: 0.04s
Fold:1, Epoch:500, train_loss: 0.2006, validation_loss: 0.2066, cost_time: 0.04s
Fold: 1, AUC_train: 0.9363, AUC_val: 0.9133, F1-score_train: 0.9254, F1-score_val: 0.8956
Fold:2, Epoch:100, train_loss: 0.1813, validation_loss: 0.1555, cost_time: 0.04s
Fold:2, Epoch:200, train_loss: 0.1671, validation_loss: 0.1449, cost_time: 0.04s
Fold:2, Epoch:300, train_loss: 0.1540, validation_loss: 0.1444, cost_time: 0.04s
Fold:2, Epoch:400, train_loss: 0.1513, validation_loss: 0.1384, cost_time: 0.04s
Fold:2, Epoch:500, train_loss: 0.1426, validation_loss: 0.1379, cost_time: 0.04s
Fold: 2, AUC_train: 0.9496, AUC_val: 0.9395, F1-score_train: 0.9401, F1-score_val: 0.9269
Fold:3, Epoch:100, train_loss: 0.1356, validation_loss: 0.1419, cost_time: 0.04s
Fold:3, Epoch:200, train_loss: 0.1219, validation_loss: 0.1384, cost_time: 0.04s
Fold:3, Epoch:300, train_loss: 0.1206, validation_loss: 0.1357, cost_time: 0.04s
Fold:3, Epoch:400, train_loss: 0.1152, validation_loss: 0.1400, cost_time: 0.04s
Fold:3, Epoch:500, train_loss: 0.1113, validation_loss: 0.1395, cost_time: 0.04s
Fold: 3, AUC_train: 0.9625, AUC_val: 0.9550, F1-score_train: 0.9535, F1-score_val: 0.9434
Fold:4, Epoch:100, train_loss: 0.1075, validation_loss: 0.1185, cost_time: 0.04s
Fold:4, Epoch:200, train_loss: 0.1155, validation_loss: 0.1250, cost_time: 0.04s
Fold:4, Epoch:300, train_loss: 0.1081, validation_loss: 0.1238, cost_time: 0.04s
Fold:4, Epoch:400, train_loss: 0.1056, validation_loss: 0.1283, cost_time: 0.04s
Fold:4, Epoch:500, train_loss: 0.0957, validation_loss: 0.1289, cost_time: 0.04s
Fold: 4, AUC_train: 0.9702, AUC_val: 0.9518, F1-score_train: 0.9629, F1-score_val: 0.9386
Fold:5, Epoch:100, train_loss: 0.1064, validation_loss: 0.0951, cost_time: 0.04s
Fold:5, Epoch:200, train_loss: 0.0983, validation_loss: 0.0978, cost_time: 0.04s
Fold:5, Epoch:300, train_loss: 0.1028, validation_loss: 0.1055, cost_time: 0.05s
Fold:5, Epoch:400, train_loss: 0.0954, validation_loss: 0.1065, cost_time: 0.04s
Fold:5, Epoch:500, train_loss: 0.0935, validation_loss: 0.1073, cost_time: 0.04s
Fold: 5, AUC_train: 0.9709, AUC_val: 0.9547, F1-score_train: 0.9647, F1-score_val: 0.9455
pred_test.shape = (1000, 5)
与LightGBM预测不同的样本数: 423
0 -1
2 -1
4 -1
8 1
16 -1
..
985 -1
987 -1
994 -1
995 -1
999 -1
Name: label, Length: 423, dtype: int64
k折交叉验证不断训练同一个模型,虽然模型最终的表现结果还可以(F1-score_val上去了),但集成各fold(各时期)的模型的结果表现依然糟糕,与baseline —— lightGBM相差甚远,都不用提交就知道分数会很低(0.63左右)了。
说明这网络模型不行啊!
可能原因:
- 集成时被早期表现较差的模型所拖累;
- 模型本身对表格数据拟合能力不够;
- 模型过拟合;
# 不用k折交叉验证,一个模型用到底
hidden = 64
activation = 'tanh'
# activation = 'tanh'
crit = nn.CrossEntropyLoss()
batch_size = 128
max_epochs = 2000
l_rate = 5e-3
dropout = 0.1
n_splits = 5
model_NN2 = NET(X_train2.shape[1], hidden, out_dim=2, activation=activation)
classifier_NN2 = NN(model_NN2, crit=crit, batch_size=batch_size, l_rate=l_rate,
max_epochs=max_epochs)
# result_SKFold_NN = SKFold(pd.DataFrame(X_train2), pd.DataFrame(Y_train2),
# pd.DataFrame(X_test2), classifier_NN2, n_splits=5)
classifier_NN2.fit(X_train2, Y_train2)
result_NN = classifier_NN2.predict(X_test2)
# c = result_LightGBM['label'] - result_SKFold_NN['label']
c = result_LightGBM['label'] - result_NN
count = 0
for i in c:
if i != 0:
count += 1
print('与LightGBM预测不同的样本数: ', count)
print(c[c!=0])
Epoch:100, train_loss: 0.2839, cost_time: 0.09s
Epoch:200, train_loss: 0.2275, cost_time: 0.09s
Epoch:300, train_loss: 0.2072, cost_time: 0.09s
Epoch:400, train_loss: 0.2097, cost_time: 0.09s
Epoch:500, train_loss: 0.1854, cost_time: 0.09s
Epoch:600, train_loss: 0.1906, cost_time: 0.09s
Epoch:700, train_loss: 0.1772, cost_time: 0.09s
Epoch:800, train_loss: 0.1749, cost_time: 0.09s
Epoch:900, train_loss: 0.1726, cost_time: 0.08s
Epoch:1000, train_loss: 0.1707, cost_time: 0.08s
Epoch:1100, train_loss: 0.1692, cost_time: 0.09s
Epoch:1200, train_loss: 0.1575, cost_time: 0.09s
Epoch:1300, train_loss: 0.1615, cost_time: 0.09s
Epoch:1400, train_loss: 0.1548, cost_time: 0.09s
Epoch:1500, train_loss: 0.1611, cost_time: 0.09s
Epoch:1600, train_loss: 0.1617, cost_time: 0.09s
Epoch:1700, train_loss: 0.1535, cost_time: 0.09s
Epoch:1800, train_loss: 0.1650, cost_time: 0.09s
Epoch:1900, train_loss: 0.1609, cost_time: 0.08s
Epoch:2000, train_loss: 0.1631, cost_time: 0.09s
与LightGBM预测不同的样本数: 424
2 -1.0
4 -1.0
6 -1.0
7 -1.0
8 1.0
...
986 1.0
988 -1.0
995 -1.0
997 -1.0
999 -1.0
Name: label, Length: 424, dtype: float64
上面这个结果说明这个神经网络模型学习遇到了瓶颈,很难再提升了。要么改模型,要么改训练方法(k折交叉训练重复训练同一个模型有提升,但提升有限,表现依然扑街),要么改数据。
4.9 SVM
表现不行。
Ref:
[1] Python3《机器学习实战》学习笔记(八):支持向量机原理篇之手撕线性SVM
from sklearn.svm import SVC
model_SVM = SVC(C=10) #C越大,对误分类的惩罚越大。
result_SKFold_SVM= SKFold(pd.DataFrame(X_train2), pd.DataFrame(Y_train2),
pd.DataFrame(X_test2), model_SVM, n_splits=5)
diff_SVM = evaluate(result_LightGBM, result_SKFold_SVM)
Fold: 1, AUC_train: 0.9001, AUC_val: 0.8614, F1-score_train: 0.8819, F1-score_val: 0.8311
Fold: 2, AUC_train: 0.8987, AUC_val: 0.8737, F1-score_train: 0.8797, F1-score_val: 0.8489
Fold: 3, AUC_train: 0.8942, AUC_val: 0.8916, F1-score_train: 0.8744, F1-score_val: 0.8711
Fold: 4, AUC_train: 0.8901, AUC_val: 0.8840, F1-score_train: 0.8688, F1-score_val: 0.8614
Fold: 5, AUC_train: 0.8945, AUC_val: 0.8836, F1-score_train: 0.8747, F1-score_val: 0.8602
与LightGBM预测不同的样本数: 271
2 -1
8 1
16 -1
23 -1
33 -1
..
983 -1
985 -1
993 1
995 -1
999 -1
Name: label, Length: 271, dtype: int64
4.10 sklearn神经网络
from sklearn.neural_network import MLPClassifier
model_MLP = MLPClassifier(hidden_layer_sizes=128, activation='relu')
result_SKFold_MLP = SKFold(pd.DataFrame(X_train2), pd.DataFrame(Y_train2),
pd.DataFrame(X_test2), model_MLP, n_splits=5)
c = result_LightGBM['label'] - result_SKFold_MLP['label']
count = 0
for i in c:
if i != 0:
count += 1
print('与LightGBM预测不同的样本数: ', count)
print(c[c!=0])
Fold: 1, AUC_train: 0.8780, AUC_val: 0.8394, F1-score_train: 0.8550, F1-score_val: 0.8045
Fold: 2, AUC_train: 0.8789, AUC_val: 0.8746, F1-score_train: 0.8558, F1-score_val: 0.8512
Fold: 3, AUC_train: 0.8838, AUC_val: 0.8828, F1-score_train: 0.8617, F1-score_val: 0.8599
Fold: 4, AUC_train: 0.8869, AUC_val: 0.8959, F1-score_train: 0.8637, F1-score_val: 0.8747
Fold: 5, AUC_train: 0.8867, AUC_val: 0.8820, F1-score_train: 0.8650, F1-score_val: 0.8579
与LightGBM预测不同的样本数: 238
2 -1
8 1
16 -1
23 -1
33 -1
..
979 -1
983 -1
993 1
995 -1
999 -1
Name: label, Length: 238, dtype: int64
神经网络在这个任务上的表现扑街了。。
5. 总结思考
这次比赛最好的分数为stacking的0.96577,再往上提升一点变得非常困难,继续提升一点分数需要耗费巨量时间和精力,投入产出比划不来,就没有继续去改进了。但从这次比赛也学到了许多,掌握了许多树模型的使用方法,以及特征工程的一点技巧。
在表格数据上,不得不说还是树模型表现更好,计算快,对算力的需求没有神经网络那么大,结果也非常棒。相反,神经网络在这个数据上表现明显不如树模型,或许是因为我采用的模型过于简单了。
透过现象看本质,这个比赛本质就是个简单的二分类问题,那么有没有一种可能,推荐系统里的DeepFM、DCN等网络模型也能用于这个问题呢?
另外,这个比赛我有点过于注重模型部分了,特征工程没有怎么去做。数据决定了你能达到的上限,模型只是帮你接近这个上限。
哈哈哈,有空再试了。欢迎各位大佬在评论区留言赐教,一起变得更强!
参考资料:
[1] Datawhale_如何打一个数据挖掘比赛V2.1
[2] 讯飞官方参考解析
[3] Kaggle上分技巧——单模K折交叉验证训练+多模型融合