如何只用bert夺冠之对比学习代码解读
有监督对比学习:Supervised Contrastive Learning:
https://zhuanlan.zhihu.com/p/136332151
1. 自监督对比学习
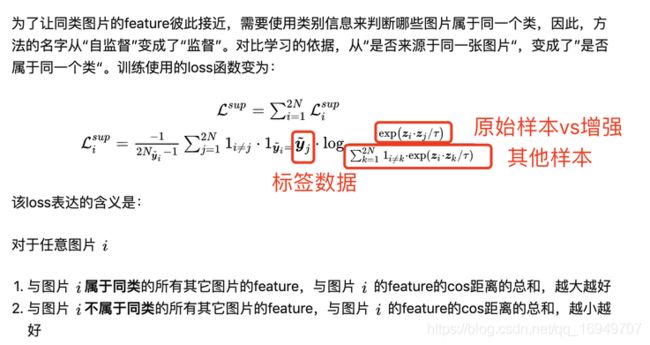
一句话总结:不使用label数据,通过数据增强构造样本,使特征提取器提取的特征在增强样本和原始样本的距离更近,其他数据特征与原始样本的更远来训练特征提取器的方法。
关键思想:一个batch=n的数据,增强一次变成2n的数据,loss函数如下:

2. 监督对比学习
把标签数据加进来,但是计算loss还是以一个batch=n,自监督计算loss的思想来做。
问:真的有用吗?
3. 代码实现
- 核心代码总结
- 将label作为输入,当然结合了bert的原始输入,另外预测的时候,还是通过嫁接一个最基础的分类model来进行预测和输出。
训练的时候,没有指定loss函数,只是在train_model中以监督对比loss和交叉熵loss最为输出,并继承与Loss类并指定了add_metric(loss),这样可能就可以了,反正也是给定一个数据,使loss最小 - 优缺点分析:
一个batch里面,正负样本就那么点,并且正样本之间的相似度不一定要高啊,在这个任务场景下,所以对比训练这个方式提升不大吧。
可能在语义相似度计算任务上,真正的语义相似度计算而不是问答任务上,效果可能会好点吧。 - 代码链接:https://github.com/xv44586/ccf_2020_qa_match/blob/main/pair-supervised-contrastive-learning.py
- 将label作为输入,当然结合了bert的原始输入,另外预测的时候,还是通过嫁接一个最基础的分类model来进行预测和输出。
class SupervisedContrastiveLearning(Loss):
"""https://arxiv.org/pdf/2011.01403.pdf"""
def __init__(self, alpha=1., T=1., **kwargs):
super(SupervisedContrastiveLearning, self).__init__(**kwargs)
self.alpha = alpha # loss weight
self.T = T # Temperature
def compute_loss(self, inputs, mask=None):
loss = self.compute_loss_of_scl(inputs)
loss = loss * self.alpha
self.add_metric(loss, name='scl_loss')
return loss
def get_label_mask(self, y_true):
"""获取batch内相同label样本"""
label = K.cast(y_true, 'int32') # 转换数据类型
label_2 = K.reshape(label, (1, -1)) # reshape成一行
mask = K.equal(label_2, label) # 这两个shape都不一样,出来的是啥?知道了,应该原来是一列,现在换成一行,所以可以比较每个位置的label是否一样了
mask = K.cast(mask, K.floatx()) # 又把它转成float类型,这样lable相等的位置为1.0,不相等的时候为0.0
mask = mask * (1 - K.eye(K.shape(y_true)[0])) # 排除对角线,即 i == j,对角线的位置的值全设置为0
return mask
def compute_loss_of_scl(self, inputs, mask=None):
y_pred, y_true = inputs
label_mask = self.get_label_mask(y_true) # mask是个二维矩阵,告诉i,j位置的lable是否一样
y_pred = K.l2_normalize(y_pred, axis=1) # 特征向量归一化
similarities = K.dot(y_pred, K.transpose(y_pred)) # 相似矩阵,相当于是点乘
similarities = similarities - K.eye(K.shape(y_pred)[0]) * 1e12 # 排除对角线,即 i == j,点乘然后排出对角线的位置
similarities = similarities / self.T # Temperature scale
similarities = K.exp(similarities) # exp
sum_similarities = K.sum(similarities, axis=-1, keepdims=True) # sum i != k, 求和得到的应该是分母
scl = similarities / sum_similarities # 这里算的还是全部的作为分子,但是我们只要把i和j位置label相同的作为分子,所以还要乘以mask
scl = K.log((scl + K.epsilon())) # sum log,取log
scl = -K.sum(scl * label_mask, axis=1, keepdims=True) / (K.sum(label_mask, axis=1, keepdims=True) + K.epsilon()) # 乘以mask
return K.mean(scl)
class CrossEntropy(Loss):
def compute_loss(self, inputs, mask=None):
pred, ytrue = inputs
ytrue = K.cast(ytrue, K.floatx())
loss = K.binary_crossentropy(ytrue, pred)
loss = K.mean(loss)
self.add_metric(loss, name='clf_loss')
return loss
# 加载预训练模型
bert = build_transformer_model(
config_path=config_path,
checkpoint_path=checkpoint_path,
model='nezha',
keep_tokens=keep_tokens,
num_hidden_layers=12,
)
# 将label作为输入
y_in = Input(shape=(None,))
output = Lambda(lambda x: x[:, 0])(bert.output)
# output相当于是特征,监督对比函数相当于是利用了label类别信息求了个loss
scl_output = SupervisedContrastiveLearning(alpha=0.1, T=0.2, output_idx=0)([output, y_in])
# scl_output是监督对比函数的loss
output = Dropout(0.1)(output)
# 这个是分类的概率
clf_output = Dense(1, activation='sigmoid')(output)
# 这个是分类的loss
clf = CrossEntropy(0)([clf_output, y_in])
# clf是分类的loss
# model模型还是以bert作为输入,分类的概率clf_output作为输出
model = keras.models.Model(bert.input, clf_output)
model.summary()
# train_model模型是用于训练的模型,bert+label作为输入,scl_output是监督对比函数的loss与clf是分类的loss作为输出
# 将loss函数作为输出,后面complile就不用指定loss函数了吗?还有这种操作,那为啥要两个loss函数啊?
train_model = keras.models.Model(bert.input + [y_in], [scl_output, clf])
optimizer = extend_with_weight_decay(Adam)
optimizer = extend_with_piecewise_linear_lr(optimizer)
opt = optimizer(learning_rate=1e-5, weight_decay_rate=0.1, exclude_from_weight_decay=['Norm', 'bias'],
lr_schedule={int(len(train_generator) * 0.1 * epochs): 1, len(train_generator) * epochs: 0}
)
train_model.compile(
optimizer=opt,
# 但是这里没有指定loss啊,所以loss是啥,所以
)
# 正常的分类模型是这么做的,在compile里面指定了loss函数
"""
model = keras.models.Model(bert.input, output)
model.summary()
model.compile(
# 指定了loss函数
loss=K.binary_crossentropy,
optimizer=Adam(2e-5), # 用足够小的学习率
metrics=['accuracy'],
)
class Evaluator(keras.callbacks.Callback):
"""评估与保存
"""
def __init__(self):
self.best_val_f1 = 0.
def on_epoch_end(self, epoch, logs=None):
val_f1 = evaluate(valid_generator)
if val_f1 > self.best_val_f1:
self.best_val_f1 = val_f1
model.save_weights('best_parimatch_model.weights')
print(
u'val_f1: %.5f, best_val_f1: %.5f\n' %
(val_f1, self.best_val_f1)
)
evaluator = Evaluator()
model.fit_generator(
train_generator.generator(),
steps_per_epoch=len(train_generator),
epochs=5,
callbacks=[evaluator],
)
"""
def evaluate(data):
P, R, TP = 0., 0., 0.
for x, _ in tqdm(data):
x_true = x[:2]
y_true = x[-1]
y_pred = model.predict(x_true)[:, 0]
y_pred = np.round(y_pred)
y_true = y_true[:, 0]
R += y_pred.sum()
P += y_true.sum()
TP += ((y_pred + y_true) > 1).sum()
print(P, R, TP)
pre = TP / R
rec = TP / P
return 2 * (pre * rec) / (pre + rec)
class Evaluator(keras.callbacks.Callback):
"""评估与保存
"""
def __init__(self, save_path):
self.best_val_f1 = 0.
self.save_path = save_path
def on_epoch_end(self, epoch, logs=None):
val_f1 = evaluate(valid_generator)
if val_f1 > self.best_val_f1:
self.best_val_f1 = val_f1
model.save_weights(self.save_path)
print(
u'val_f1: %.5f, best_val_f1: %.5f\n' %
(val_f1, self.best_val_f1)
)
def predict_to_file(path='pair_submission.tsv'):
preds = []
for x, _ in tqdm(test_generator):
x = x[:2]
pred = model.predict(x).argmax(axis=1)
# pred = np.round(pred)
pred = pred.astype(int)
preds.append(pred)
preds = np.concatenate(preds)
ret = []
for d, p in zip(test_data, preds):
q_id, _, r_id, _, _ = d
ret.append([str(q_id), str(r_id), str(p)])
with open(path, 'w', encoding='utf8') as f:
for l in ret:
f.write('\t'.join(l) + '\n')
if __name__ == '__main__':
save_path = 'best_pair_scl_model.weights'
evaluator = Evaluator(save_path)
train_model.fit_generator(
train_generator.generator(),
steps_per_epoch=len(train_generator),
epochs=epochs,
callbacks=[evaluator],
)
model.load_weights(save_path)
predict_to_file('pair_scl.tsv')