scikit-learn(sklearn)学习笔记十 支持向量机
书接上次笔记,我们通过把二维的数据映射到三维,再用超平面进行划分。但是这也是有很大的问题的,维数越高越难以计算。于是在上次笔记的最后,采用了更换核函数来满足支持向量机的分类要求。
klearn在skearn中可选择以下几种选项
- linear 线性核,解决问题为线性。
- poly 多项式核,解决问题为偏线性。
- sigmoid 双曲正切核,解决问题为非线性。
- rbf 高斯径向基,解决偏为非线性。
所以要研究kernel应该如何选取
from sklearn.datasets import load_breast_cancer
from sklearn.svm import SVC
from sklearn.model_selection import train_test_split
import matplotlib.pyplot as plt
import numpy as np
from time import time
import datetime
data=load_breast_cancer()
X=data.data
y=data.target
#X.shape
#np.unique(y)
plt.scatter(X[:,0],X[:,1],c=y)
plt.show()
Xtrain,Xtest,Ytrain,Ytest=train_test_split(X,y,test_size=0.3,random_state=420)
Kernel=["linear","poly","rbf","sigmoid"]
for kernel in Kernel:
time0=time()
clf=SVC(kernel=kernel,gamma="auto",cache_size=5000).fit(Xtrain,Ytrain)
print(kernel,clf.score(Xtest,Ytest))
print(datetime.datetime.fromtimestamp(time()-time0).strftime("%M:%S:%f"))
linear 0.9298245614035088
00:00:505116
因为poly 多项式核还有一个参数degree默认的degree等于3,这让我电脑运行了很久都没有出结果,如果我把poly去掉。
linear 0.9298245614035088
00:00:487102
rbf 0.5964912280701754
00:00:053013
sigmoid 0.5964912280701754
00:00:005007
如果我将ploy中的degree改成等于三
linear 0.9298245614035088
00:00:491111
poly 0.9239766081871345
00:00:091014
rbf 0.5964912280701754
00:00:052018
sigmoid 0.5964912280701754
00:00:006002
那为什么看起来万能的rbf,他所提供模型的误差会比线性模型相差很多,我们先查看一下我们所用的数据集
data=pd.DataFrame(X)
data.describe([0.01,0.05,0.1,0.25,0.5,0.75,0.9,0.99]).T
count mean std ... 90% 99% max
0 569.0 14.127292 3.524049 ... 19.530000 24.371600 28.11000
1 569.0 19.289649 4.301036 ... 24.992000 30.652000 39.28000
2 569.0 91.969033 24.298981 ... 129.100000 165.724000 188.50000
3 569.0 654.889104 351.914129 ... 1177.400000 1786.600000 2501.00000
4 569.0 0.096360 0.014064 ... 0.114820 0.132888 0.16340
5 569.0 0.104341 0.052813 ... 0.175460 0.277192 0.34540
6 569.0 0.088799 0.079720 ... 0.203040 0.351688 0.42680
7 569.0 0.048919 0.038803 ... 0.100420 0.164208 0.20120
8 569.0 0.181162 0.027414 ... 0.214940 0.259564 0.30400
9 569.0 0.062798 0.007060 ... 0.072266 0.085438 0.09744
10 569.0 0.405172 0.277313 ... 0.748880 1.291320 2.87300
11 569.0 1.216853 0.551648 ... 1.909400 2.915440 4.88500
12 569.0 2.866059 2.021855 ... 5.123200 9.690040 21.98000
13 569.0 40.337079 45.491006 ... 91.314000 177.684000 542.20000
14 569.0 0.007041 0.003003 ... 0.010410 0.017258 0.03113
15 569.0 0.025478 0.017908 ... 0.047602 0.089872 0.13540
16 569.0 0.031894 0.030186 ... 0.058520 0.122292 0.39600
17 569.0 0.011796 0.006170 ... 0.018688 0.031194 0.05279
18 569.0 0.020542 0.008266 ... 0.030120 0.052208 0.07895
19 569.0 0.003795 0.002646 ... 0.006185 0.012650 0.02984
20 569.0 16.269190 4.833242 ... 23.682000 30.762800 36.04000
21 569.0 25.677223 6.146258 ... 33.646000 41.802400 49.54000
22 569.0 107.261213 33.602542 ... 157.740000 208.304000 251.20000
23 569.0 880.583128 569.356993 ... 1673.000000 2918.160000 4254.00000
24 569.0 0.132369 0.022832 ... 0.161480 0.188908 0.22260
25 569.0 0.254265 0.157336 ... 0.447840 0.778644 1.05800
26 569.0 0.272188 0.208624 ... 0.571320 0.902380 1.25200
27 569.0 0.114606 0.065732 ... 0.208940 0.269216 0.29100
28 569.0 0.290076 0.061867 ... 0.360080 0.486908 0.66380
29 569.0 0.083946 0.018061 ... 0.106320 0.140628 0.20750
[30 rows x 13 columns]
通过观察平均值和方差,我们有理由怀疑量纲不是同一,数据的分布是偏态的,所以我们进行一下数据的标准化。
from sklearn.preprocessing import StandardScaler
X=StandardScaler().fit_transform(X)
我们再用这个数据集进行运算
linear 0.9766081871345029
00:00:008999
poly 0.9649122807017544
00:00:004002
rbf 0.9707602339181286
00:00:008001
sigmoid 0.9532163742690059
00:00:004001
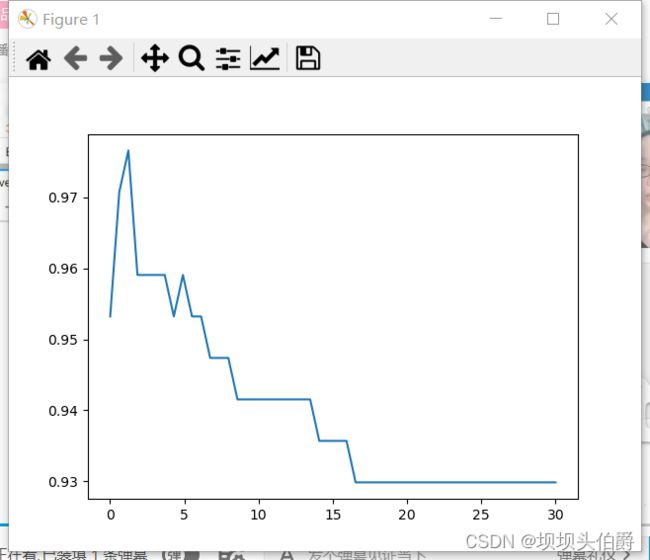
我们可以发现线性核函数和高斯核函数的效果都十分的好,我们应该作何选择呢,线性核函数是没有参数的没有可以进行调整,而高斯核函数有gamma值可以调整,所以我们来画高斯核函数的学习曲线。
score=[]
gamma_range=np.logspace(-10,1,50)
for i in gamma_range:
clf=SVC(kernel="rbf",gamma=i,cache_size=5000).fit(Xtrain,Ytrain)
score.append(clf.score(Xtest,Ytest))
print(max(score),gamma_range[score.index(max(score))])
plt.plot(gamma_range,score)
plt.show()
0.9766081871345029 0.012067926406393264
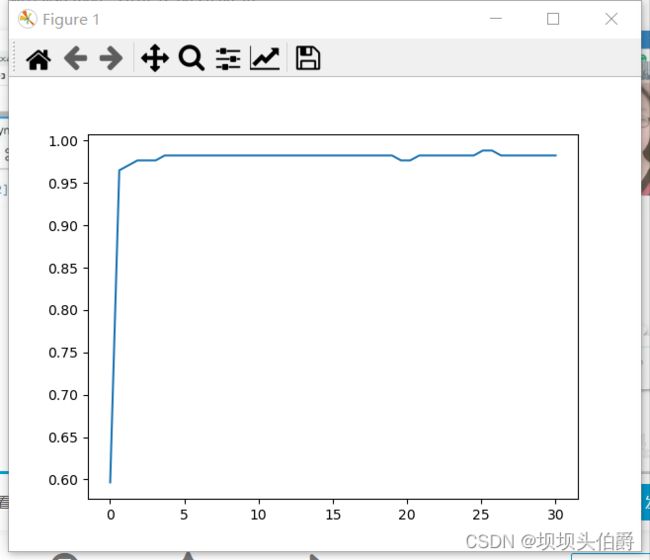
 我们一直假设训练样本在样本空间或特征空间食线性可分的,即存在一个超平面能将不同类的样本完全划分开。然而,在现实任务中往往很难确定合适的核函数使得训练样本在特征空间中线性可分;退一步说,即便恰好找到了某个核函数使训练样本在特征空间中线性可分,也很难断定这个貌似线性可分的结果不是由于过拟合造成的。缓解该问题的一个方法是允许支持向量机在一些样本上出错,为此要引入“软间隔”的概念。
我们一直假设训练样本在样本空间或特征空间食线性可分的,即存在一个超平面能将不同类的样本完全划分开。然而,在现实任务中往往很难确定合适的核函数使得训练样本在特征空间中线性可分;退一步说,即便恰好找到了某个核函数使训练样本在特征空间中线性可分,也很难断定这个貌似线性可分的结果不是由于过拟合造成的。缓解该问题的一个方法是允许支持向量机在一些样本上出错,为此要引入“软间隔”的概念。
score=[]
C_range=np.linspace(0.01,30,50)
for i in C_range:
clf=SVC(kernel="linear",C=i,cache_size=5000).fit(Xtrain,Ytrain)
score.append(clf.score(Xtest,Ytest))
print(max(score),C_range[score.index(max(score))])
plt.plot(C_range,score)
plt.show()
0.9766081871345029 1.2340816326530613
score=[]
C_range=np.linspace(0.01,30,50)
for i in C_range:
clf=SVC(kernel="rbf",C=i,cache_size=5000).fit(Xtrain,Ytrain)
score.append(clf.score(Xtest,Ytest))
print(max(score),C_range[score.index(max(score))])
plt.plot(C_range,score)
plt.show()
0.9883040935672515 25.103673469387758