回归,分类评价指标及案例
模型评估指标(RMSE、MSE、MAE、R2准确率、召回率、F1、ROC曲线、AUC曲线、PR曲线)
1、回归模型评估指标
a、RMSE(Root Mean Square Error)均方根误差
衡量观测值与真实值之间的偏差。常用来作为机器学习模型预测结果衡量的标准。

b、MSE(Mean Square Error)均方误差
通过平方的形式便于求导,所以常被用作线性回归的损失函数。用了MSE为代价函数的模型因为要最小化这个异常值带来的误差,就会尽量贴近异常值,也就是对outliers(异常值)赋予更大的权重。这样就会影响总体的模型效果。

c、MAE(Mean Absolute Error)平均绝对误差
是绝对误差的平均值。可以更好地反映预测值误差的实际情况,相比MSE来说,MAE在数据里有不利于预测结果异常值的情况下
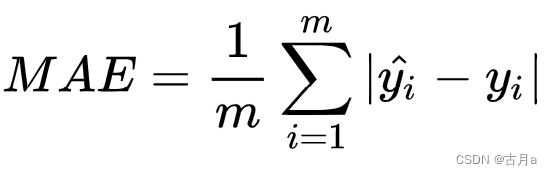
d、SD(Standard Deviation)标准差
方差的算术平均根。用于衡量一组数值的离散程度。

R2(R- Square)拟合优度
R2=SSR/SST=1-SSE/SST
其中:SST=SSR+SSE,


Error = Bias + Variance
Error反映的是整个模型的准确度,Bias反映的是模型在样本上的输出与真实值之间的误差,即模型本身的精准度,Variance反映的是模型每一次输出结果与模型输出期望之间的误差,即模型的稳定性.
2、分类
对数损失不适用于样本不均衡时的分类评估指标
ROC-AUC可作为样本正负不均衡时的分类评估指标
如果我们想让少数情况被正确预测,就用ROC-AUC作为评估指标
F1- Score和PR曲线在正样本极少时适用于作为分类评估指标
F1- Score和PR曲线在FP比FN更重要时,适用于作为分类评估指标
1.准确率(accuracy)
所有预测正确的样本/总的样本 = (TP+TN)/总
from sklearn.metrics import accuracy
accuracy = accuracy_score(y_test, y_predict)
2.查准率(precision)
预测为正的样本中有多少是真的正样本。两种可能,一种就是把正类预测为正类(TP),另一种就是把负类预测为正类(FP)
![]()
from sklearn.metrics import precision_score
precision = precision_score(y_test, y_predict)
3.查全率/召回率(recall)
样本中的正样本有多少被预测正确了。两种可能,一种是把原来的正类预测成正类(TP),另一种就是把原来的正类预测为负类(FN):
from sklearn.metrics import recall_score
recall = recall_score(y_test, y_predict)
#recall得到的是一个list,是每一类的召回率
from sklearn.metrics import f1_score
f1_score(y_test, y_predict)
5.PR曲线
PR曲线是准确率和召回率的点连成的线。
**PR曲线与ROC曲线的相同点是都采用了TPR (Recall),都可以用AUC来衡量分类器的效果。不同点是ROC曲线使用了FPR,而PR曲线使用了Precision,
因此PR曲线的两个指标都聚焦于正例。类别不平衡问题中由于主要关心正例,所以在此情况下PR曲线被广泛认为优于ROC曲线。
6.ROC(Receiver Operating Characteristic)曲线,又称接受者操作特征曲线
通过动态地调整截断点,从最高的得分开始(实际上是从正无穷开始,对应着ROC曲线的零点),逐渐调整到最低得分,每一个截断点都会对应一个FPR和TPR,在ROC图上绘制出每个截断点对应的位置再连接所有点就得到最终的ROC曲线。
ROC的含义为概率曲线,AUC的含义为正负类可正确分类的程度。
TPR(True Positive Rate)真正例率/查准率P
真实的正例中,被预测为正例的比例:TPR = TP/(TP+FN)。
FPR(False Positive Rate)假正例率****
真实的反例中,被预测为正例的比例:FPR = FP/(TN+FP)。
理想分类器TPR=1,FPR=0。ROC曲线越接近左上角,代表模型越好,即ACU接近1
截断点thresholds
指的就是区分正负预测结果的阈值
7.AUC
计算:分别随机从正负样本集中抽取一个正样本,一个负样本,正样本的预测值大于负样本的概率。
AUC = 1,是完美分类器,采用这个预测模型时,不管设定什么阈值都能得出完美预测。绝大多数预测的场合,不存在完美分类器。0.5 < AUC < 1,优于随机猜测。AUC = 0.5,跟随机猜测一样。AUC < 0.5,比随机猜测还差。
eg1.Binary-class classification
import numpy as np
np.random.seed(10)
import matplotlib.pyplot as plt
from sklearn.datasets import make_classification
from sklearn.preprocessing import label_binarize
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import train_test_split
from sklearn.metrics import roc_curve
X, y = make_classification(n_samples=80000)
# print(X[0], y[0])
# (80000, 20) (80000,)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.5)
X_train, X_train_lr, y_train, y_train_lr = train_test_split(X_train, y_train, test_size=0.5)
from keras.models import Sequential
from keras.layers import Dense
from sklearn.metrics import auc
model = Sequential()
model.add(Dense(20, input_dim=20, activation='relu'))
model.add(Dense(40, activation='relu'))
model.add(Dense(1, activation='sigmoid'))
model.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy'])
model.fit(X_train, y_train, epochs=5, batch_size=100, verbose=1)
y_pred = model.predict(X_test).ravel()
print(y_pred.shape)
fpr, tpr, thresholds = roc_curve(y_test, y_pred)
roc_auc = auc(fpr, tpr)
plt.figure(1)
plt.plot([0, 1], [0, 1], 'k--')
plt.plot(fpr, tpr, label='Keras (area = {:.3f})'.format(roc_auc))
plt.xlabel('False positive rate')
plt.ylabel('True positive rate')
plt.title('ROC curve')
plt.legend(loc='best')
plt.show()
# Zoom in view of the upper left corner.
plt.figure(2)
plt.xlim(0, 0.2)
plt.ylim(0.8, 1)
plt.plot([0, 1], [0, 1], 'k--')
plt.plot(fpr, tpr, label='Keras (area = {:.3f})'.format(roc_auc))
plt.xlabel('False positive rate')
plt.ylabel('True positive rate')
plt.title('ROC curve (zoomed in at top left)')
plt.legend(loc='best')
plt.show()
# (Optional) Prediction probability density function(PDF)
import numpy as np
from scipy.interpolate import UnivariateSpline
from matplotlib import pyplot as plt
def plot_pdf(y_pred, y_test, name=None, smooth=500):
positives = y_pred[y_test == 1]
negatives = y_pred[y_test == 0]
N = positives.shape[0]
n = N//smooth
s = positives
p, x = np.histogram(s, bins=n) # bin it into n = N//10 bins
x = x[:-1] + (x[1] - x[0])/2 # convert bin edges to centers
f = UnivariateSpline(x, p, s=n)
plt.plot(x, f(x))
N = negatives.shape[0]
n = N//smooth
s = negatives
p, x = np.histogram(s, bins=n) # bin it into n = N//10 bins
x = x[:-1] + (x[1] - x[0])/2 # convert bin edges to centers
f = UnivariateSpline(x, p, s=n)
plt.plot(x, f(x))
plt.xlim([0.0, 1.0])
plt.xlabel('density')
plt.ylabel('density')
plt.title('PDF-{}'.format(name))
plt.show()
plot_pdf(y_pred, y_test, 'Keras')
宏平均(Macro-averaging)和微平均(Micro-averaging):
用途:用于多个类别的分类
宏平均:是先对每一个类统计指标值,然后在对所有类求算术平均值。
微平均:是对数据集中的每一个实例不分类别进行统计建立全局混淆矩阵,然后计算相应指标
eg2.Multi-class classification
from sklearn.datasets import make_classification
from sklearn.preprocessing import label_binarize
from keras.models import Sequential
from keras.layers import Dense
import numpy as np
from scipy import interp
import matplotlib.pyplot as plt
from itertools import cycle
from sklearn.model_selection import train_test_split
from sklearn.metrics import roc_curve, auc
# 标签共三类
n_classes = 3
X, y = make_classification(n_samples=80000, n_features=20, n_informative=3, n_redundant=0, n_classes=n_classes,
n_clusters_per_class=2)
# print(X.shape, y.shape)
# print(X[0], y[0])
# (80000, 20) (80000,)
# [-1.90920853 -1.30052757 -0.76903467 -3.2546519 -0.02947816 0.14105006
# 0.43556031 -0.81300607 -0.94553296 -0.92774495 1.49041451 -0.4443121
# -1.16342165 -0.32997815 -1.02907045 -0.39950447 -0.711287 0.51382424
# 2.88822258 -2.0935274 ]
# 1
# Binarize the output相当于one_hot
y = label_binarize(y, classes=[0, 1, 2])
# print(y.shape, y[0])
# (80000, 3) [0 1 0]
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.5)
model = Sequential()
model.add(Dense(20, input_dim=20, activation='relu'))
model.add(Dense(40, activation='relu'))
model.add(Dense(3, activation='softmax'))
model.compile(loss='categorical_crossentropy', optimizer='adam', metrics=['accuracy'])
model.fit(X_train, y_train, epochs=1, batch_size=100, verbose=1)
y_pred = model.predict(X_test)
# print(y_pred.shape)
# (40000, 3)
# Compute ROC curve and ROC area for each class
fpr = dict()
tpr = dict()
roc_auc = dict()
for i in range(n_classes):
# scores = np.array([0.1, 0.4, 0.35, 0.8])
# fpr, tpr, thresholds = metrics.roc_curve(y, scores, pos_label=2)
# y 就是标准值,scores 是每个预测值对应的阳性概率,比如0.1就是指第一个数预测为阳性的概率为0.1,很显然,
# y 和 socres应该有相同多的元素,都等于样本数。pos_label=2 是指在y中标签为2的是标准阳性标签,其余值是阴性。
# 接下来选取一个阈值计算TPR/FPR,阈值的选取规则是在scores值中从大到小的以此选取,于是第一个选取的阈值是0.8
# label=[1,1,2,2] scores=[0.1,0.4,0.35,0.8] thresholds=[0.8,0.4,0.35,0.1] 以threshold为0.8为例,将0.8与
# scores 中所有值比较大小得到预测值,[0,0,0,1].对于label中两个1,其概率分别为0.1,0.4,小于阈值0.8,判定为
# 负样本,而他们的label是1,说明他们确实是负样本,判断正确,是两个TN;两个2,对应概率为0.35,0.8,0.35小于
# 0.8,判定为负样本,但是label是2,应该是个正样本,所以这是个FN;最后0.8>=0.8,这是个TP,所以最后的结果是
# :1个TP,2个TN,1个FN,0个FP
fpr[i], tpr[i], thresholds = roc_curve(y_test[:, i], y_pred[:, i]) # (40000,)
# print(fpr[i].shape)# (5491,)# (6562,)# (4271,)
roc_auc[i] = auc(fpr[i], tpr[i])
# 计算microROC曲线和ROC面积
# .ravel()将多维数组转换为一维数组
fpr["micro"], tpr["micro"] , thresholds = roc_curve(y_test.ravel(), y_pred.ravel()) # (120000,)
roc_auc["micro"] = auc(fpr["micro"], tpr["micro"])
# 计算macroROC曲线和ROC面积
# 首先,汇总所有的假阳性率
# np.unique() 该函数是去除数组中的重复数字,并进行排序之后输出。
# print(np.concatenate([fpr[i] for i in range(n_classes)]).shape) (16324,)
all_fpr = np.unique(np.concatenate([fpr[i] for i in range(n_classes)])) # (7901,)
# 然后插值所有的ROC曲线在这一点
# np.zeros_like() 这个函数的意思就是生成一个和你所给数组a相同shape的全0数组。
mean_tpr = np.zeros_like(all_fpr)
for i in range(n_classes):
mean_tpr += interp(all_fpr, fpr[i], tpr[i])
# 最后求平均值并计算AUC
mean_tpr /= n_classes
fpr["macro"] = all_fpr
tpr["macro"] = mean_tpr
roc_auc["macro"] = auc(fpr["macro"], tpr["macro"])
# Plot all ROC curves
plt.figure(1)
plt.plot(fpr["micro"], tpr["micro"], color='deeppink', linestyle=':', linewidth=4,
label='micro-average ROC curve (area = {0:0.2f})'.format(roc_auc["micro"]))
plt.plot(fpr["macro"], tpr["macro"],color='navy', linestyle=':', linewidth=4,
label='macro-average ROC curve (area = {0:0.2f})'.format(roc_auc["macro"]))
colors = cycle(['aqua', 'darkorange', 'cornflowerblue'])
for i, color in zip(range(n_classes), colors):
plt.plot(fpr[i], tpr[i], color=color, linewidth=2,
label='ROC curve of class {0} (area = {1:0.2f})'.format(i, roc_auc[i]))
plt.plot([0, 1], [0, 1], 'k--', linewidth=2)
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.05])
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.title('Some extension of Receiver Operating Characteristic to multi-class')
plt.legend(loc='best')
plt.show()
# Zoom in view of the upper left corner.
plt.figure(2)
plt.xlim(0, 0.2)
plt.ylim(0.8, 1)
plt.plot(fpr["micro"], tpr["micro"],color='deeppink', linestyle=':', linewidth=4,
label='micro-average ROC curve (area = {0:0.2f})'.format(roc_auc["micro"]))
plt.plot(fpr["macro"], tpr["macro"],color='navy', linestyle=':', linewidth=4,
label='macro-average ROC curve (area = {0:0.2f})'.format(roc_auc["macro"]))
colors = cycle(['aqua', 'darkorange', 'cornflowerblue'])
for i, color in zip(range(n_classes), colors):
plt.plot(fpr[i], tpr[i], color=color, linewidth=2,
label='ROC curve of class {0} (area = {1:0.2f})'.format(i, roc_auc[i]))
plt.plot([0, 1], [0, 1], 'k--', linewidth=2)
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.title('ROC curve (zoomed in at top left)')
plt.legend(loc='best')
plt.show()
混淆矩阵
def plot_confusion_matrix(title, y_true, y_pred, labels):
import matplotlib.pyplot as plt
from sklearn.metrics import confusion_matrix
cm = confusion_matrix(y_true, y_pred)
# np.newaxis的作用就是在这一位置增加一个一维,这一位置指的是np.newaxis所在的位置,比较抽象,需要配合例子理解。
# x1 = np.array([1, 2, 3, 4, 5])
# the shape of x1 is (5,)
# x1_new = x1[:, np.newaxis]
# now, the shape of x1_new is (5, 1)
cm_normalized = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]
# print (cm, '\n\n', cm_normalized)
# [[1 0 0 0 0]
# [0 1 0 0 0]
# [0 0 1 0 0]
# [0 0 0 1 0]
# [0 0 0 0 1]]
# [[1. 0. 0. 0. 0.]
# [0. 1. 0. 0. 0.]
# [0. 0. 1. 0. 0.]
# [0. 0. 0. 1. 0.]
# [0. 0. 0. 0. 1.]]
tick_marks = np.array(range(len(labels))) + 0.5
# [0.5 1.5 2.5 3.5 4.5 5.5]
np.set_printoptions(precision=2)
plt.figure(figsize=(10, 8), dpi=120)
ind_array = np.arange(len(labels))
x, y = np.meshgrid(ind_array, ind_array)
# print(ind_array, '\n\n', x, '\n\n', y)
# [0 1 2 3 4 5]
# [[0 1 2 3 4 5]
# [0 1 2 3 4 5]
# [0 1 2 3 4 5]
# [0 1 2 3 4 5]
# [0 1 2 3 4 5]
# [0 1 2 3 4 5]]
# [[0 0 0 0 0 0]
# [1 1 1 1 1 1]
# [2 2 2 2 2 2]
# [3 3 3 3 3 3]
# [4 4 4 4 4 4]
# [5 5 5 5 5 5]]
intFlag = 0 # 标记在图片中对文字是整数型还是浮点型
for x_val, y_val in zip(x.flatten(), y.flatten()):
# plt.text()函数用于设置文字说明。
if (intFlag):
c = cm[y_val][x_val]
plt.text(x_val, y_val, "%d" % (c,), color='red', fontsize=8, va='center', ha='center')
else:
c = cm_normalized[y_val][x_val]
if (c > 0.01):
plt.text(x_val, y_val, "%0.2f" % (c,), color='red', fontsize=7, va='center', ha='center')
else:
plt.text(x_val, y_val, "%d" % (0,), color='red', fontsize=7, va='center', ha='center')
cmap = plt.cm.binary
if(intFlag):
plt.imshow(cm, interpolation='nearest', cmap=cmap)
else:
plt.imshow(cm_normalized, interpolation='nearest', cmap=cmap)
plt.gca().set_xticks(tick_marks, minor=True)
plt.gca().set_yticks(tick_marks, minor=True)
plt.gca().xaxis.set_ticks_position('none')
plt.gca().yaxis.set_ticks_position('none')
plt.grid(True, which='minor', linestyle='-')
plt.gcf().subplots_adjust(bottom=0.15)
plt.title(title)
plt.colorbar()
xlocations = np.array(range(len(labels)))
plt.xticks(xlocations, labels, rotation=90)
plt.yticks(xlocations, labels)
plt.ylabel('Index of True Classes')
plt.xlabel('Index of Predict Classes')
plt.savefig('confusion_matrix.jpg', dpi=300)
plt.show()
title='Confusion Matrix'
labels = ['A', 'B', 'C', 'F', 'G']
y_true = [1, 2, 3, 4, 5]# np.loadtxt(r'/home/dingtom/a.txt')
y_pred = [1, 2, 3, 4, 5]# np.loadtxt(r'/home/dingtom/b.txt')
plot_confusion_matrix(title, y_true,y_pred, labels)
