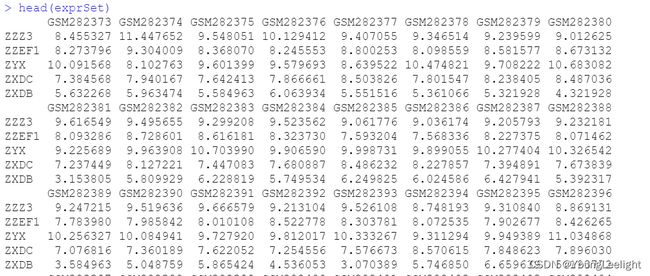
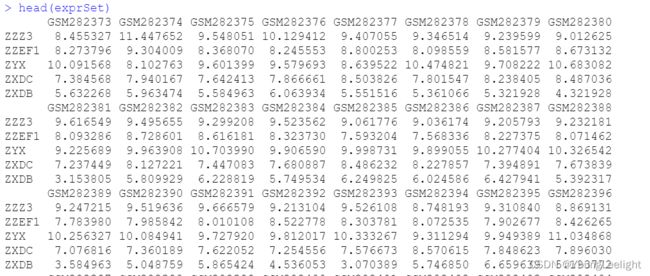
Surv单因素批量生存分析使用 cox批量生存回归分析有点像deseq2的design差异分析designG:\r\2021_1203_geo\GEO-master\GSE11121_survival
批量生存分析
input
codes
output
input: 表达矩阵 和 meta 信息
表达矩阵
meta信息
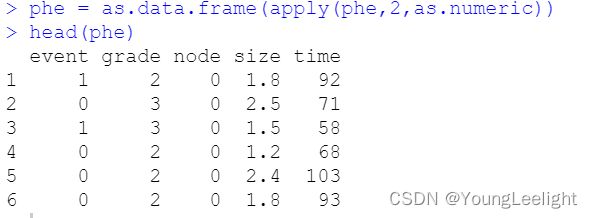
phe = as.data.frame(apply(phe,2,as.numeric))
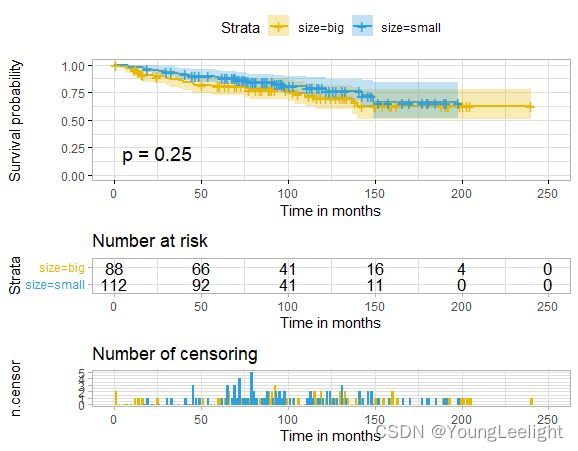
phe$size=ifelse(phe$size>median(phe$size),'big','small')
head(phe)
存活分析
library(survival)
library(survminer)
# 利用ggsurvplot快速绘制漂亮的生存曲线图
sfit <- survfit(data=phe,Surv(time, event)~size)
sfit
summary(sfit)
ggsurvplot(sfit, conf.int=F, pval=TRUE)
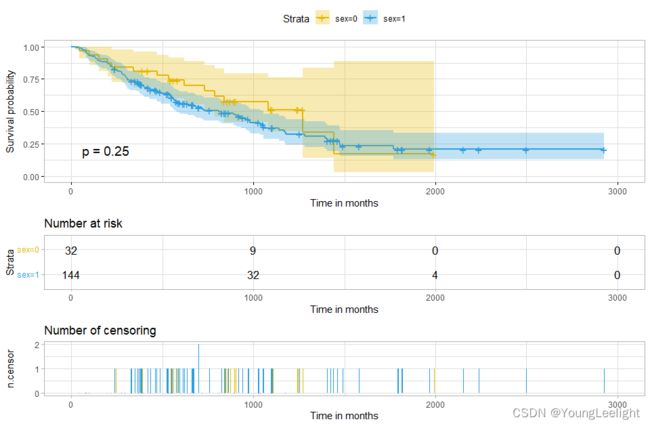
ggsurvplot(sfit,palette = c("#E7B800", "#2E9FDF"),
risk.table =TRUE,pval =TRUE,
conf.int =TRUE,xlab ="Time in months",
ggtheme =theme_light(),
ncensor.plot = TRUE)
挑选感兴趣的基因做差异分析
phe$CBX4=ifelse(exprSet['CBX4',]>median(exprSet['CBX4',]),'high','low')
head(phe)
table(phe$CBX4)
ggsurvplot(survfit(Surv(time, event)~CBX4, data=phe), conf.int=F, pval=TRUE)
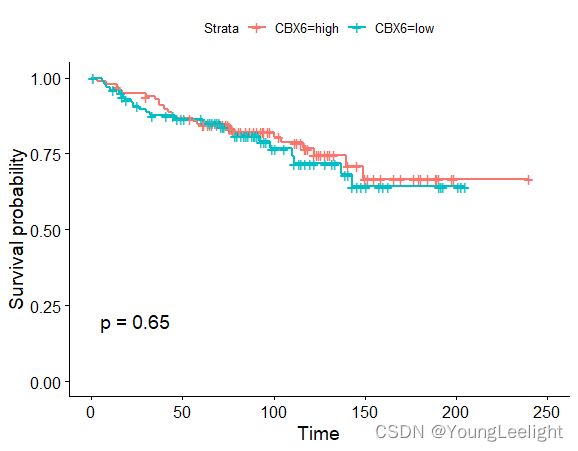
phe$CBX6=ifelse(exprSet['CBX6',]>median(exprSet['CBX6',]),'high','low')
head(phe)
table(phe$CBX6)
ggsurvplot(survfit(Surv(time, event)~CBX6, data=phe), conf.int=F, pval=TRUE)
with()R语言中的函数用于通过评估函数参数内的表达式来修改 DataFrame 的数据。
用法: with(x, expr)
参数:
x: DataFrame
expr:修改数据的表达式
# R program to modify data of an object
# Creating a data frame
df = data.frame(
"Name" = c("abc", "def", "ghi"),
"Language" = c("R", "Python", "Java"),
"Age" = c(22, 25, 45)
)
df
# Calling with() function
with(df, {df$Age <- df$Age + 10; print(df$Age)})
##批量
批量生存分析 使用 logrank test 方法
mySurv=with(phe,Surv(time, event))
log_rank_p <- apply(exprSet , 1 , function(gene){#求得一个基因高低表达 生存分析的p值
#gene=exprSet[1,]
phe$group=ifelse(gene>median(gene),'high','low')
data.survdiff=survdiff(mySurv~group,data=phe)
p.val = 1 - pchisq(data.survdiff$chisq, length(data.survdiff$n) - 1)
return(p.val)
})
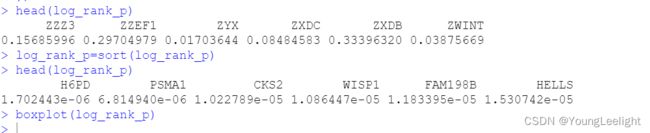
log_rank_p=sort(log_rank_p)
head(log_rank_p)
boxplot(log_rank_p)
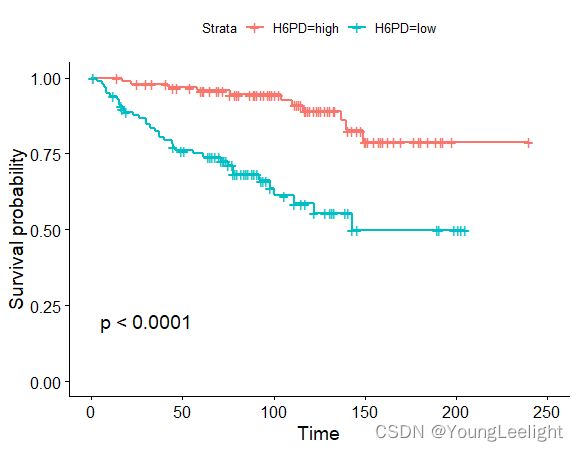
phe$H6PD=ifelse(exprSet['H6PD',]>median(exprSet['H6PD',]),'high','low')
table(phe$H6PD)
ggsurvplot(survfit(Surv(time, event)~H6PD, data=phe), conf.int=F, pval=TRUE)
head(log_rank_p)
ZZZ3 ZZEF1 ZYX ZXDC ZXDB ZWINT
0.15685996 0.29704979 0.01703644 0.08484583 0.33396320 0.03875669
log_rank_p=sort(log_rank_p)
head(log_rank_p)
boxplot(log_rank_p)
可以找到p值最小的为 HDP6基因
boxplot(log_rank_p)
phe$H6PD=ifelse(exprSet['H6PD',]>median(exprSet['H6PD',]),'high','low')
table(phe$H6PD)
ggsurvplot(survfit(Surv(time, event)~H6PD, data=phe), conf.int=F, pval=TRUE)
前面如果我们使用了WGCNA找到了跟grade相关的基因模块,然后在这里就可以跟生存分析的显著性基因做交集
###这样就可以得到既能跟grade相关,又有临床预后意义的基因啦。
批量生存分析 使用 coxph 回归方法 因为cox可以进行多因素回归分析
colnames(phe)
head(phe)
mySurv=with(phe,Surv(time, event))
cox_results <-apply(exprSet , 1 , function(gene){ #把每个基因 分为高低表达两组 进行多因素回归分析
group=ifelse(gene>median(gene),'high','low')
survival_dat <- data.frame(group=group,
grade=phe$grade,
size=phe$size,stringsAsFactors = F)
m=coxph(mySurv ~ grade + size + group,
data = survival_dat)
beta <- coef(m)
se <- sqrt(diag(vcov(m)))
HR <- exp(beta) #提取风险因子
HRse <- HR * se
#summary(m)
tmp <- round(cbind(coef = beta, se = se, z = beta/se, p = 1 - pchisq((beta/se)^2, 1),
HR = HR, HRse = HRse,
HRz = (HR - 1) / HRse, HRp = 1 - pchisq(((HR - 1)/HRse)^2, 1),
HRCILL = exp(beta - qnorm(.975, 0, 1) * se),
HRCIUL = exp(beta + qnorm(.975, 0, 1) * se)), 3)
return(tmp['grouplow',])
})
cox_results=t(cox_results)
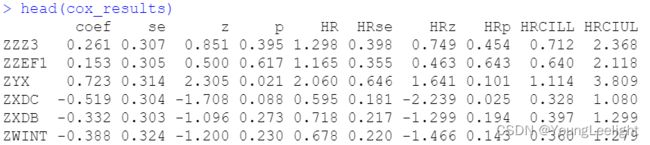
head(cox_results)
table(cox_results[,4]<0.05)
cox_results[cox_results[,4]<0.05,]
length(setdiff(rownames(cox_results[cox_results[,4]<0.05,]),
names(log_rank_p[log_rank_p<0.05])
))
length(setdiff( names(log_rank_p[log_rank_p<0.05]),
rownames(cox_results[cox_results[,4]<0.05,])
))
length(unique( names(log_rank_p[log_rank_p<0.05]),
rownames(cox_results[cox_results[,4]<0.05,])
))
save(log_rank_p,cox_results,exprSet,phe,file = 'surviva.Rdata')
多因素回归分析之后 可以找到所有符合满意p值的基因
https://zhuanlan.zhihu.com/p/164351709
要将单变量coxph函数一次应用于多个协变量,请输入以下命令:单基因批量cox回归分析 单因素批量cox回归分析
要将单变量coxph函数一次应用于多个协变量,请输入以下命令:
covariates <- c("age", "sex", "ph.karno", "ph.ecog", "wt.loss")
univ_formulas <- sapply(covariates,
function(x) as.formula(paste('Surv(time, status)~', x)))
univ_models <- lapply( univ_formulas, function(x){coxph(x, data = lung)})
# 提取数据,并制作数据表格
univ_results <- lapply(univ_models,
function(x){
x <- summary(x)
p.value<-signif(x$wald["pvalue"], digits=2)
wald.test<-signif(x$wald["test"], digits=2)
beta<-signif(x$coef[1], digits=2);#coeficient beta
HR <-signif(x$coef[2], digits=2);#exp(beta)
HR.confint.lower <- signif(x$conf.int[,"lower .95"], 2)
HR.confint.upper <- signif(x$conf.int[,"upper .95"],2)
HR <- paste0(HR, " (",
HR.confint.lower, "-", HR.confint.upper, ")")
res<-c(beta, HR, wald.test, p.value)
names(res)<-c("beta", "HR (95% CI for HR)", "wald.test",
"p.value")
return(res)
#return(exp(cbind(coef(x),confint(x))))
})
res <- t(as.data.frame(univ_results, check.names = FALSE))
as.data.frame(res)
beta HR (95% CI for HR) wald.test p.value
age 0.019 1 (1-1) 4.1 0.042
sex -0.53 0.59 (0.42-0.82) 10 0.0015
ph.karno -0.016 0.98 (0.97-1) 7.9 0.005
ph.ecog 0.48 1.6 (1.3-2) 18 2.7e-05
wt.loss 0.0013 1 (0.99-1) 0.05 0.83