linkET,环境因子相关性 + mantel test 可视化新选择
文章目录
- 前言
- linkET 安装
- 官方示例
-
-
- 数据预处理
- 热图
- Mantel test
- 网络
-
- 总结
前言
环境因子相关性 + mantel test 组合图,网上的可视化教程都是用R package:ggcor,但是好像没人维护了,安装起来也有一些小问题……
最近发现有另一个package:linkET,同一个作者,熟悉的味道,这里简单安利一下。
下面的内容都可以从大佬的Github上找到:https://github.com/Hy4m/linkET
linkET 安装
# install.packages("devtools")
devtools::install_github("Hy4m/linkET", force = TRUE)
packageVersion("linkET")
官方示例
数据预处理
library(linkET)
matrix_data(list(mtcars = mtcars))
#> A matrix data object:
#> Number: 1
#> Names: mtcars
#> Dimensions: 32 rows, 11 columns
#> Row names: Mazda RX4, Mazda RX4 Wag, Datsun 710, Hornet 4 Drive, Hor...
#> Column names: mpg, cyl, disp, hp, drat, wt, qsec, vs, am, gear, carb
matrix_data(list(mtcars = mtcars)) %>%
as_md_tbl()
#> # A tibble: 352 × 3
#> .rownames .colnames mtcars
#> *
#> 1 Mazda RX4 mpg 21
#> 2 Mazda RX4 Wag mpg 21
#> 3 Datsun 710 mpg 22.8
#> 4 Hornet 4 Drive mpg 21.4
#> 5 Hornet Sportabout mpg 18.7
#> # … with 342 more rows
as_matrix_data(mtcars)
#> A matrix data object:
#> Number: 1
#> Names: mtcars
#> Dimensions: 32 rows, 11 columns
#> Row names: Mazda RX4, Mazda RX4 Wag, Datsun 710, Hornet 4 Drive, Hor...
#> Column names: mpg, cyl, disp, hp, drat, wt, qsec, vs, am, gear, carb
as_md_tbl(mtcars)
#> # A tibble: 352 × 3
#> .rownames .colnames mtcars
#> *
#> 1 Mazda RX4 mpg 21
#> 2 Mazda RX4 Wag mpg 21
#> 3 Datsun 710 mpg 22.8
#> 4 Hornet 4 Drive mpg 21.4
#> 5 Hornet Sportabout mpg 18.7
#> # … with 342 more rows
## special function for correlation matrix
correlate(mtcars) %>%
as_matrix_data()
#> A matrix data object:
#> Number: 4
#> Names: r, p, lower_ci, upper_ci
#> Dimensions: 11 rows, 11 columns
#> Row names: mpg, cyl, disp, hp, drat, wt, qsec, vs, am, gear, carb
#> Column names: mpg, cyl, disp, hp, drat, wt, qsec, vs, am, gear, carb
correlate(mtcars) %>%
as_md_tbl()
#> # A tibble: 121 × 6
#> .rownames .colnames r p lower_ci upper_ci
#> *
#> 1 mpg mpg 1 0 1 1
#> 2 cyl mpg -0.852 6.11e-10 -0.926 -0.716
#> 3 disp mpg -0.848 9.38e-10 -0.923 -0.708
#> 4 hp mpg -0.776 1.79e- 7 -0.885 -0.586
#> 5 drat mpg 0.681 1.78e- 5 0.436 0.832
#> # … with 111 more rows
热图
代码如下(示例):
correlate(mtcars) %>%
as_md_tbl() %>%
qcorrplot() +
geom_square()
library(vegan)
#> 载入需要的程辑包:permute
#> 载入需要的程辑包:lattice
#> This is vegan 2.5-7
data("varespec")
data("varechem")
correlate(varespec[1:30], varechem) %>%
qcorrplot() +
geom_square() +
scale_fill_gradientn(colours = RColorBrewer::brewer.pal(11, "RdBu"))
Mantel test
library(dplyr)
#>
#> 载入程辑包:'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
data("varechem", package = "vegan")
data("varespec", package = "vegan")
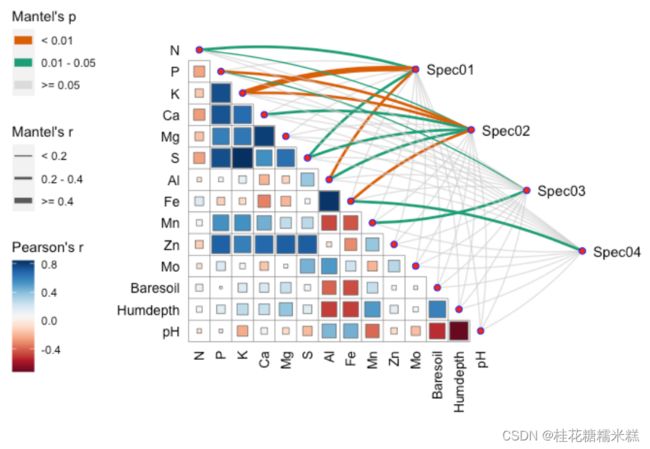
mantel <- mantel_test(varespec, varechem,
spec_select = list(Spec01 = 1:7,
Spec02 = 8:18,
Spec03 = 19:37,
Spec04 = 38:44)) %>%
mutate(rd = cut(r, breaks = c(-Inf, 0.2, 0.4, Inf),
labels = c("< 0.2", "0.2 - 0.4", ">= 0.4")),
pd = cut(p, breaks = c(-Inf, 0.01, 0.05, Inf),
labels = c("< 0.01", "0.01 - 0.05", ">= 0.05")))
#> `mantel_test()` using 'bray' dist method for 'spec'.
#> `mantel_test()` using 'euclidean' dist method for 'env'.
qcorrplot(correlate(varechem), type = "lower", diag = FALSE) +
geom_square() +
geom_couple(aes(colour = pd, size = rd),
data = mantel,
curvature = nice_curvature()) +
scale_fill_gradientn(colours = RColorBrewer::brewer.pal(11, "RdBu")) +
scale_size_manual(values = c(0.5, 1, 2)) +
scale_colour_manual(values = color_pal(3)) +
guides(size = guide_legend(title = "Mantel's r",
override.aes = list(colour = "grey35"),
order = 2),
colour = guide_legend(title = "Mantel's p",
override.aes = list(size = 3),
order = 1),
fill = guide_colorbar(title = "Pearson's r", order = 3))
网络
correlate(mtcars) %>%
as.igraph() %>%
plot(layout = layout_with_circular)
总结
在强调一下,以上,在linkET的GitHub上都与可以看到,这里只是做了搬运……linkET