Seurat 对象的构建和信息提取
文章目录
-
- 从 CellRanger 输出构建
- 从 h5 文件构建
- 从表达矩阵构建
- read10x函数报错,手动根据三个文件构建表达矩阵
- Crea
- 批量构建seurat 对象
- 对Seurat对象的理解和信息提取
- dgCMatrix
目前构建Seurat对象有以下几种方法:
- 从 CellRanger 输出构建
- 从 h5 文件构建
- 从表达矩阵构建
从 CellRanger 输出构建
公司在完成表达定量后,通常会使用 CellRanger 对数据进行简单的分析,得到以下三个文件。
- barcodes.tsv 用于储存细胞信息
- genes.tsv 用于储存基因信息
- matrix.mtx 表达矩阵
##更新内容:
Cellranger >= 3.0版本后 gene.tsv 变成了 features.tsv.gz
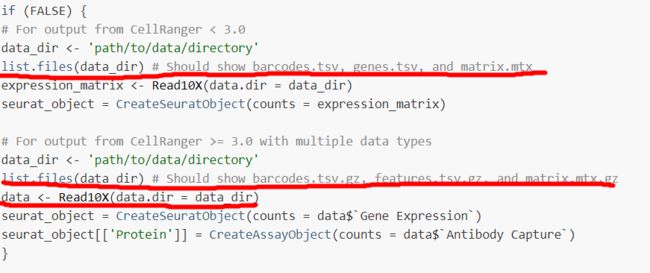
ScRNAdata = Read10X(data.dir = "GSE134809_RAW/")
# GSE134809_RAW/ 文件夹下面包含了上述三个文件
# 此时的 ScRNAdata 是一个稀疏矩阵 dgCMatrix
# > class(ScRNAdata)
# [1] "dgCMatrix"
# attr(,"package")
# [1] "Matrix"
# 构建 Seurat 对象
# 初步过滤一般不需要修改参数,除非数据实在太难看
Seurat_object <- CreateSeuratObject(
counts = ScRNAdata, # 表达矩阵,可以为稀疏矩阵,也可以为普通矩阵
min.cells = 3, # 去除在小于3个细胞中表达的基因
min.features = 200) # 去除只有 200 个以下基因表达的细胞
从 h5 文件构建
ScRNAdata <- Read10X_h5(filename = "GSM3489182_Donor_01_raw_gene_bc_matrices_h5.h5")
Seurat_object <- CreateSeuratObject(
counts = ScRNAdata,
min.cells = 3,
min.features = 200)
从表达矩阵构建
ScRNA_exp <- read.table("data/GSM2829942/GSM2829942_HE6W_LA.TPM.txt",row.names = 1,header = T)
Seurat_object <- CreateSeuratObject(
counts = ScRNA_exp,
min.cells = 3,
min.features = 200)
read10x函数报错,手动根据三个文件构建表达矩阵
library(Matrix)
matrix_dir = "/opt/sample345/outs/filtered_feature_bc_matrix/"
barcode.path <- paste0(matrix_dir, "barcodes.tsv.gz")
features.path <- paste0(matrix_dir, "features.tsv.gz")
matrix.path <- paste0(matrix_dir, "matrix.mtx.gz")
mat <- readMM(file = matrix.path)
feature.names = read.delim(features.path,
header = FALSE,
stringsAsFactors = FALSE)
barcode.names = read.delim(barcode.path,
header = FALSE,
stringsAsFactors = FALSE)
colnames(mat) = barcode.names$V1
rownames(mat) = feature.names$V1
Seurat_object <- CreateSeuratObject(
counts = mat,
min.cells = 3,
min.features = 200)
Crea
teSeuratObject其他参数
- counts :表达矩阵 cells as columns and features as rows
- project : Project name for the Seurat object
- assay : Name of the initial assay,数据transform后产生一个新的assay,以示区别
- names.field :表达矩阵的colname是cell name,If your cells are named as BARCODE_CLUSTER_CELLTYPE in
the input matrix, set names.field to 3 to set the initial identities to CELLTYPE - names.delim : If your cells are named as BARCODE-CLUSTER-CELLTYPE, set this to “-” to separate
the cell name into its component parts for picking the relevant field. - min.cells : 至少多少个cell表达这个feature,不然该feature不会保留
- min.features :每个cell 至少表达多少个feature,不然不会保留
- row.names :When counts is a data.frame or data.frame-derived object: an optional vector of feature names to be used
批量构建seurat 对象
# load data from 10X and create Seurat object
path <- '/public/home/djs/huiyu/2020_NC_ESCC'
folders <- list.files(path, pattern = "YX")
sceList = lapply(folders,function(folder){
CreateSeuratObject(counts = Read10X(folder),
project = folder, min.cells = 3,
min.features = 200)
})
# load data from counts and create Seurat object
path <- '/public/home/djs/huiyu/2020_NC_ESCC'
files <- list.files(path, pattern = "YX")
sceList = lapply(files,function(file){
counts <- read.table(file,header=T)
CreateSeuratObject(counts = counts),
project = file, min.cells = 3,
min.features = 200)
})
对Seurat对象的理解和信息提取
> ?seurat
Slots:
‘raw.data’ The raw project data
‘data’ The normalized expression matrix (log-scale)
‘scale.data’ scaled (default is z-scoring each gene) expression
matrix; used for dimensional reduction and heatmap
visualization
‘var.genes’ Vector of genes exhibiting high variance across single
cells
‘is.expr’ Expression threshold to determine if a gene is expressed
(0 by default)
‘ident’ THe 'identity class' for each cell
‘meta.data’ Contains meta-information about each cell, starting
with number of genes detected (nFeature) and the original
identity class (orig.ident); more information is added using
‘AddMetaData’
‘project.name’ Name of the project (for record keeping)
‘dr’ List of stored dimensional reductions; named by technique
‘assay’ List of additional assays for multimodal analysis; named
by technique
‘hvg.info’ The output of the mean/variability analysis for all
genes
‘imputed’ Matrix of imputed gene scores
‘cell.names’ Names of all single cells (column names of the
expression matrix)
‘cluster.tree’ List where the first element is a phylo object
containing the phylogenetic tree relating different identity
classes
‘snn’ Spare matrix object representation of the SNN graph
‘calc.params’ Named list to store all calculation-related
parameter choices
‘kmeans’ Stores output of gene-based clustering from ‘DoKMeans’
‘spatial’ Stores internal data and calculations for spatial
mapping of single cells
‘misc’ Miscellaneous spot to store any data alongside the object
(for example, gene lists)
‘version’ Version of package used in object creation
>sceList1.integrated@ ## tab自动补全会出来一些可调用对象信息
sceList1.integrated@assays sceList1.integrated@active.ident sceList1.integrated@reductions sceList1.integrated@misc sceList1.integrated@tools
sceList1.integrated@meta.data sceList1.integrated@graphs sceList1.integrated@images sceList1.integrated@version
sceList1.integrated@active.assay sceList1.integrated@neighbors sceList1.integrated@project.name sceList1.integrated@commands
>sceList1.integrated@assays
$RNA
Assay data with 22695 features for 51225 cells
Top 10 variable features:
IGKC, JCHAIN, IGHA1, DCD, S100A7, S100A9, IGHG4, S100A8, SCGB2A2,
CXCL10
$integrated
Assay data with 2000 features for 51225 cells
Top 10 variable features:
KRT2, S100A7, S100A8, S100A9, CCL19, CFD, COL1A1, CALML5, PTGDS, G0S2
> sceList1.integrated$
sceList1.integrated$orig.ident sceList1.integrated$nCount_RNA sceList1.integrated$nFeature_RNA sceList1.integrated$percent.mt
# seurat对象中的@符号和$ 符号的区别:
# 它们是从两个不同的面向对象系统中提取变量的符号
> str(sceList1.integrated)
Formal class 'Seurat' [package "SeuratObject"] with 13 slots
..@ assays :List of 2
.. ..$ RNA :Formal class 'Assay' [package "SeuratObject"] with 8 slots
.. .. .. ..@ counts :Formal class 'dgCMatrix' [package "Matrix"] with 6 slots
.. .. .. .. .. ..@ i : int [1:103511748] 1 15 26 48 68 72 82 83 95 107 ...
.. .. .. .. .. ..@ p : int [1:51226] 0 1188 3784 6674 8887 10773 11874 14671 18297 19910 ...
.. .. .. .. .. ..@ Dim : int [1:2] 22695 51225
.. .. .. .. .. ..@ Dimnames:List of 2
.. .. .. .. .. .. ..$ : chr [1:22695] "RP11-34P13.7" "FO538757.2" "AP006222.2" "RP4-669L17.10" ...
.. .. .. .. .. .. ..$ : chr [1:51225] "AAACCTGAGTATCGAA-1_1" "AAACCTGCAAGCCATT-1_1" "AAACCTGCAGGTGGAT-1_1" "AAACCTGGTGTTGAGG-1_1" ...
.. .. .. .. .. ..@ x : num [1:103511748] 1 1 1 1 1 1 3 5 1 1 ...
.. .. .. .. .. ..@ factors : list()
.. .. .. ..@ data :Formal class 'dgCMatrix' [package "Matrix"] with 6 slots
.. .. .. .. .. ..@ i : int [1:103511748] 1 15 26 48 68 72 82 83 95 107 ...
.. .. .. .. .. ..@ p : int [1:51226] 0 1188 3784 6674 8887 10773 11874 14671 18297 19910 ...
.. .. .. .. .. ..@ Dim : int [1:2] 22695 51225
.. .. .. .. .. ..@ Dimnames:List of 2
.. .. .. .. .. .. ..$ : chr [1:22695] "RP11-34P13.7" "FO538757.2" "AP006222.2" "RP4-669L17.10" ...
.. .. .. .. .. .. ..$ : chr [1:51225] "AAACCTGAGTATCGAA-1_1" "AAACCTGCAAGCCATT-1_1" "AAACCTGCAGGTGGAT-1_1" "AAACCTGGTGTTGAGG-1_1" ...
.. .. .. .. .. ..@ x : num [1:103511748] 1.65 1.65 1.65 1.65 1.65 ...
.. .. .. .. .. ..@ factors : list()
.. .. .. ..@ scale.data : num[0 , 0 ]
.. .. .. ..@ key : chr "rna_"
.. .. .. ..@ assay.orig : NULL
.. .. .. ..@ var.features : chr [1:10000] "IGKC" "JCHAIN" "IGHA1" "DCD" ...
.. .. .. ..@ meta.features:'data.frame': 22695 obs. of 5 variables:
.. .. .. .. ..$ vst.mean : num [1:22695] 0.002987 0.198087 0.096223 0.003982 0.000273 ...
.. .. .. .. ..$ vst.variance : num [1:22695] 0.003017 0.213552 0.10516 0.003967 0.000273 ...
.. .. .. .. ..$ vst.variance.expected : num [1:22695] 0.003564 0.275453 0.122526 0.004815 0.000292 ...
.. .. .. .. ..$ vst.variance.standardized: num [1:22695] 0.847 0.775 0.858 0.824 0.936 ...
.. .. .. .. ..$ vst.variable : logi [1:22695] FALSE FALSE FALSE FALSE TRUE FALSE ...
.. .. .. ..@ misc : list()
.. ..$ integrated:Formal class 'Assay' [package "SeuratObject"] with 8 slots
.. .. .. ..@ counts :Formal class 'dgCMatrix' [package "Matrix"] with 6 slots
.. .. .. .. .. ..@ i : int(0)
.. .. .. .. .. ..@ p : int 0
.. .. .. .. .. ..@ Dim : int [1:2] 0 0
.. .. .. .. .. ..@ Dimnames:List of 2
.. .. .. .. .. .. ..$ : NULL
.. .. .. .. .. .. ..$ : NULL
.. .. .. .. .. ..@ x : num(0)
.. .. .. .. .. ..@ factors : list()
.. .. .. ..@ data :Formal class 'dgCMatrix' [package "Matrix"] with 6 slots
.. .. .. .. .. ..@ i : int [1:87809565] 0 1 2 3 4 5 6 7 8 9 ...
.. .. .. .. .. ..@ p : int [1:51226] 0 2000 4000 6000 8000 10000 12000 14000 16000 18000 ...
.. .. .. .. .. ..@ Dim : int [1:2] 2000 51225
.. .. .. .. .. ..@ Dimnames:List of 2
.. .. .. .. .. .. ..$ : chr [1:2000] "KRT2" "S100A7" "S100A8" "S100A9" ...
.. .. .. .. .. .. ..$ : chr [1:51225] "AAACCTGAGTATCGAA-1_1" "AAACCTGCAAGCCATT-1_1" "AAACCTGCAGGTGGAT-1_1" "AAACCTGGTGTTGAGG-1_1" ...
.. .. .. .. .. ..@ x : num [1:87809565] 0.1256 0.0352 0.0537 0.0382 0.0469 ...
.. .. .. .. .. ..@ factors : list()
.. .. .. ..@ scale.data : num [1:2000, 1:51225] -0.141 -0.241 -0.343 -0.282 -0.167 ...
.. .. .. .. ..- attr(*, "dimnames")=List of 2
.. .. .. .. .. ..$ : chr [1:2000] "KRT2" "S100A7" "S100A8" "S100A9" ...
.. .. .. .. .. ..$ : chr [1:51225] "AAACCTGAGTATCGAA-1_1" "AAACCTGCAAGCCATT-1_1" "AAACCTGCAGGTGGAT-1_1" "AAACCTGGTGTTGAGG-1_1" ...
.. .. .. ..@ key : chr "integrated_"
.. .. .. ..@ assay.orig : NULL
.. .. .. ..@ var.features : chr [1:2000] "KRT2" "S100A7" "S100A8" "S100A9" ...
.. .. .. ..@ meta.features:'data.frame': 2000 obs. of 0 variables
.. .. .. ..@ misc : NULL
..@ meta.data :'data.frame': 51225 obs. of 4 variables:
.. ..$ orig.ident : chr [1:51225] "H01" "H01" "H01" "H01" ...
.. ..$ nCount_RNA : num [1:51225] 2387 11240 8597 8637 7151 ...
.. ..$ nFeature_RNA: int [1:51225] 1188 2596 2890 2213 1886 1101 2797 3626 1613 1453 ...
.. ..$ percent.mt : num [1:51225] 7.75 1.92 7.68 3.39 4.32 ...
..@ active.assay: chr "integrated"
..@ active.ident: Factor w/ 15 levels "H01","H02","H03",..: 1 1 1 1 1 1 1 1 1 1 ...
.. ..- attr(*, "names")= chr [1:51225] "AAACCTGAGTATCGAA-1_1" "AAACCTGCAAGCCATT-1_1" "AAACCTGCAGGTGGAT-1_1" "AAACCTGGTGTTGAGG-1_1" ...
..@ graphs : list()
..@ neighbors : list()
..@ reductions :List of 1
.. ..$ pca:Formal class 'DimReduc' [package "SeuratObject"] with 9 slots
.. .. .. ..@ cell.embeddings : num [1:51225, 1:30] -0.0372 14.9466 -0.4031 14.318 14.3181 ...
.. .. .. .. ..- attr(*, "dimnames")=List of 2
.. .. .. .. .. ..$ : chr [1:51225] "AAACCTGAGTATCGAA-1_1" "AAACCTGCAAGCCATT-1_1" "AAACCTGCAGGTGGAT-1_1" "AAACCTGGTGTTGAGG-1_1" ...
.. .. .. .. .. ..$ : chr [1:30] "PC_1" "PC_2" "PC_3" "PC_4" ...
.. .. .. ..@ feature.loadings : num [1:2000, 1:30] 0.02892 0.01455 0.01631 0.01283 0.00121 ...
.. .. .. .. ..- attr(*, "dimnames")=List of 2
.. .. .. .. .. ..$ : chr [1:2000] "KRT2" "S100A7" "S100A8" "S100A9" ...
.. .. .. .. .. ..$ : chr [1:30] "PC_1" "PC_2" "PC_3" "PC_4" ...
.. .. .. ..@ feature.loadings.projected: num[0 , 0 ]
.. .. .. ..@ assay.used : chr "integrated"
.. .. .. ..@ global : logi FALSE
.. .. .. ..@ stdev : num [1:30] 12.49 10.65 8.12 7.59 6.39 ...
.. .. .. ..@ key : chr "PC_"
.. .. .. ..@ jackstraw :Formal class 'JackStrawData' [package "SeuratObject"] with 4 slots
.. .. .. .. .. ..@ empirical.p.values : num[0 , 0 ]
.. .. .. .. .. ..@ fake.reduction.scores : num[0 , 0 ]
.. .. .. .. .. ..@ empirical.p.values.full: num[0 , 0 ]
.. .. .. .. .. ..@ overall.p.values : num[0 , 0 ]
.. .. .. ..@ misc :List of 1
.. .. .. .. ..$ total.variance: num 1931
..@ images : list()
..@ project.name: chr "SeuratProject"
..@ misc : list()
..@ version :Classes 'package_version', 'numeric_version' hidden list of 1
.. ..$ : int [1:3] 4 1 0
..@ commands :List of 5
.. ..$ FindIntegrationAnchors :Formal class 'SeuratCommand' [package "SeuratObject"] with 5 slots
.. .. .. ..@ name : chr "FindIntegrationAnchors"
.. .. .. ..@ time.stamp : POSIXct[1:1], format: "2022-07-13 16:24:31"
.. .. .. ..@ assay.used : chr [1:15] "RNA" "RNA" "RNA" "RNA" ...
.. .. .. ..@ call.string: chr "FindIntegrationAnchors(object.list = sceList1, anchor.features = features)"
.. .. .. ..@ params :List of 15
.. .. .. .. ..$ assay : chr [1:15] "RNA" "RNA" "RNA" "RNA" ...
.. .. .. .. ..$ anchor.features : chr [1:2000] "KRT2" "S100A7" "S100A8" "S100A9" ...
.. .. .. .. ..$ scale : logi TRUE
.. .. .. .. ..$ normalization.method: chr "LogNormalize"
.. .. .. .. ..$ reduction : chr "cca"
.. .. .. .. ..$ l2.norm : logi TRUE
.. .. .. .. ..$ dims : int [1:30] 1 2 3 4 5 6 7 8 9 10 ...
.. .. .. .. ..$ k.anchor : num 5
.. .. .. .. ..$ k.filter : num 200
.. .. .. .. ..$ k.score : num 30
.. .. .. .. ..$ max.features : num 200
.. .. .. .. ..$ nn.method : chr "annoy"
.. .. .. .. ..$ n.trees : num 50
.. .. .. .. ..$ eps : num 0
.. .. .. .. ..$ verbose : logi TRUE
.. ..$ withCallingHandlers :Formal class 'SeuratCommand' [package "SeuratObject"] with 5 slots
.. .. .. ..@ name : chr "withCallingHandlers"
.. .. .. ..@ time.stamp : POSIXct[1:1], format: "2022-07-13 16:32:44"
.. .. .. ..@ assay.used : NULL
.. .. .. ..@ call.string: chr [1:2] "withCallingHandlers(expr, warning = function(w) if (inherits(w, " " classes)) tryInvokeRestart(\"muffleWarning\"))"
.. .. .. ..@ params :List of 9
.. .. .. .. ..$ new.assay.name : chr "integrated"
.. .. .. .. ..$ normalization.method: chr "LogNormalize"
.. .. .. .. ..$ features : chr [1:2000] "KRT2" "S100A7" "S100A8" "S100A9" ...
.. .. .. .. ..$ dims : int [1:30] 1 2 3 4 5 6 7 8 9 10 ...
.. .. .. .. ..$ k.weight : num 100
.. .. .. .. ..$ sd.weight : num 1
.. .. .. .. ..$ preserve.order : logi FALSE
.. .. .. .. ..$ eps : num 0
.. .. .. .. ..$ verbose : logi TRUE
.. ..$ FindVariableFeatures.RNA:Formal class 'SeuratCommand' [package "SeuratObject"] with 5 slots
.. .. .. ..@ name : chr "FindVariableFeatures.RNA"
.. .. .. ..@ time.stamp : POSIXct[1:1], format: "2022-07-14 10:23:10"
.. .. .. ..@ assay.used : chr "RNA"
.. .. .. ..@ call.string: chr [1:2] "FindVariableFeatures(sceList0.integrated, selection.method = \"vst\", " " nfeatures = 10000, assay = \"RNA\")"
.. .. .. ..@ params :List of 12
.. .. .. .. ..$ assay : chr "RNA"
.. .. .. .. ..$ selection.method : chr "vst"
.. .. .. .. ..$ loess.span : num 0.3
.. .. .. .. ..$ clip.max : chr "auto"
.. .. .. .. ..$ mean.function :function (mat, display_progress)
.. .. .. .. ..$ dispersion.function:function (mat, display_progress)
.. .. .. .. ..$ num.bin : num 20
.. .. .. .. ..$ binning.method : chr "equal_width"
.. .. .. .. ..$ nfeatures : num 10000
.. .. .. .. ..$ mean.cutoff : num [1:2] 0.1 8
.. .. .. .. ..$ dispersion.cutoff : num [1:2] 1 Inf
.. .. .. .. ..$ verbose : logi TRUE
.. ..$ ScaleData.integrated :Formal class 'SeuratCommand' [package "SeuratObject"] with 5 slots
.. .. .. ..@ name : chr "ScaleData.integrated"
.. .. .. ..@ time.stamp : POSIXct[1:1], format: "2022-07-15 10:18:30"
.. .. .. ..@ assay.used : chr "integrated"
.. .. .. ..@ call.string: chr "ScaleData(sceList0.integrated, verbose = FALSE)"
.. .. .. ..@ params :List of 10
.. .. .. .. ..$ features : chr [1:2000] "KRT2" "S100A7" "S100A8" "S100A9" ...
.. .. .. .. ..$ assay : chr "integrated"
.. .. .. .. ..$ model.use : chr "linear"
.. .. .. .. ..$ use.umi : logi FALSE
.. .. .. .. ..$ do.scale : logi TRUE
.. .. .. .. ..$ do.center : logi TRUE
.. .. .. .. ..$ scale.max : num 10
.. .. .. .. ..$ block.size : num 1000
.. .. .. .. ..$ min.cells.to.block: num 3000
.. .. .. .. ..$ verbose : logi FALSE
.. ..$ RunPCA.integrated :Formal class 'SeuratCommand' [package "SeuratObject"] with 5 slots
.. .. .. ..@ name : chr "RunPCA.integrated"
.. .. .. ..@ time.stamp : POSIXct[1:1], format: "2022-07-15 10:18:57"
.. .. .. ..@ assay.used : chr "integrated"
.. .. .. ..@ call.string: chr "RunPCA(sceList1.integrated, npcs = 30, verbose = FALSE)"
.. .. .. ..@ params :List of 10
.. .. .. .. ..$ assay : chr "integrated"
.. .. .. .. ..$ npcs : num 30
.. .. .. .. ..$ rev.pca : logi FALSE
.. .. .. .. ..$ weight.by.var : logi TRUE
.. .. .. .. ..$ verbose : logi FALSE
.. .. .. .. ..$ ndims.print : int [1:5] 1 2 3 4 5
.. .. .. .. ..$ nfeatures.print: num 30
.. .. .. .. ..$ reduction.name : chr "pca"
.. .. .. .. ..$ reduction.key : chr "PC_"
.. .. .. .. ..$ seed.use : num 42
..@ tools :List of 1
.. ..$ Integration:Formal class 'IntegrationData' [package "Seurat"] with 7 slots
.. .. .. ..@ neighbors : NULL
.. .. .. ..@ weights : NULL
.. .. .. ..@ integration.matrix: NULL
.. .. .. ..@ anchors :'data.frame': 818530 obs. of 5 variables:
.. .. .. .. ..$ cell1 : num [1:818530] 1 1 2 4 5 6 6 6 6 7 ...
.. .. .. .. ..$ cell2 : num [1:818530] 1206 2087 221 3115 1335 ...
.. .. .. .. ..$ score : num [1:818530] 0.711 0.658 0 0.368 0.105 ...
.. .. .. .. ..$ dataset1: int [1:818530] 1 1 1 1 1 1 1 1 1 1 ...
.. .. .. .. ..$ dataset2: int [1:818530] 2 2 2 2 2 2 2 2 2 2 ...
.. .. .. ..@ offsets : NULL
.. .. .. ..@ objects.ncell : NULL
.. .. .. ..@ sample.tree : num [1:14, 1:2] -4 -3 -8 1 3 4 -2 6 -11 5 ...
- 应该主要关注这一块
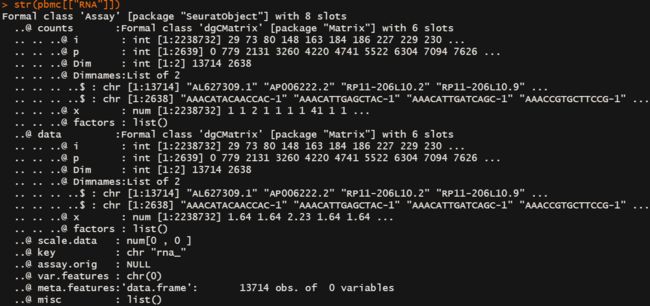
counts 储存raw data
data 储存 log transfform / scTransform后的data
scale.data 储存 缩放后的数据,可能是var.feature或者是全部基因
dgCMatrix
dgCMatrix is a class from the Matrix R package ==> 我认为Matrix是“父类”,dgCMatrix是“子类”,seurat object的数据槽是具体“实例”。
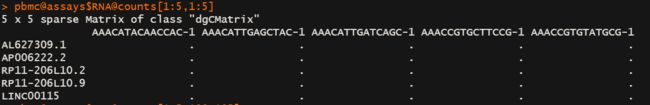
dgCMatrix是一种稀疏矩阵,具体长相如下:
有数值的是具体数值,没数值的就是个点点,可以转化为array,这些点就会变成0,而且数据大小会膨胀,注意是膨胀。
下面是他的数据组织结构
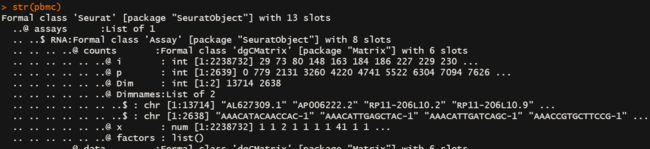
上述信息描述了::seurat object 包含了13ge slot,第一个slot称为assays,其中assays的class属性为Assay包括8个slot,有一个slot被称为counts,class属性为dgCMatrix,包含了6个slot。
counts的class属性为dgCMatrix,包含了6个slot,这里面就包含了我们的reads/UMI的信息==>raw data。
可以通过显式访问:pbmc@assays$RNA@counts
或者这样访问:pbmc[["RNA"]]@counts
-
第一个slot
名称为“i”,包括了每一个具体数值的row indes,点/0这样的数值不包括在内
i is 0-based, not 1-based like everything else in R -
第二个slot
名称为“p”,the cumulative number of data values as we move from one column to the next column, left to right. The first value is always 0。
and the length of p is one more than the number of columns. We can compute p for any matrix:c(0, cumsum(colSums(m != 0))) -
第三个slot
类似于数据框的维度,描述了稀疏矩阵的维度 dim -
第四个slot
类似于数据框,记录每个维度的名称,描述了稀疏矩阵的维度 dimname -
第五个slot
名称“x”,每个具体数值的数值大小 -
第六个slot
暂时还没搞清楚,不过看名字大概是一个描述因子向量的地方。

