R语言单因素方差分析+差异显著字母法标注+逐行详细解释
- R语言单因素方差分析
代码如下
df <- read.csv("data.csv",header = TRUE,row.names = 1)
library(reshape2)
df <- melt(df,id=c())
names(df) <- c('trt', 'val')
df
aov1 <- aov(val~trt,data=df)
summary(aov1)
library(agricolae)
data <- LSD.test(aov1,'trt',p.adj = 'bonferroni')#'bonferroni'#对P值进行修正
data
print(data$groups)
plot(data)
开始逐行解释:
导入数据
#导入数据
df <- read.csv("data.csv",header = TRUE,row.names = 1)
df
> df
a b c d e f k
1 3.186224 3.262900 2.397264 2.300343 1.806937 2.711331 2.945837
2 2.975125 3.068194 2.962235 2.233887 2.136561 4.185355 3.018140
3 3.150602 4.297190 2.518045 2.169607 2.473778 3.948050 2.785514
宽数据变为长数据,并且重命名
library(reshape2)
df <- melt(df,id=c())
names(df) <- c('trt', 'val')
df
> df
trt val
2 a 2.975125
3 a 3.150602
4 b 3.262900
5 b 3.068194
6 b 4.297190
7 c 2.397264
8 c 2.962235
9 c 2.518045
10 d 2.300343
11 d 2.233887
12 d 2.169607
13 e 1.806937
14 e 2.136561
15 e 2.473778
16 f 2.711331
17 f 4.185355
18 f 3.948050
19 k 2.945837
20 k 3.018140
21 k 2.785514
查看方差分析结果:
其中Pr(>F)=0.00661<0.05,说明组间存在差异显著
aov1 <- aov(val~trt,data=df)
summary(aov1)
> summary(aov1)
Df Sum Sq Mean Sq F value Pr(>F)
trt 6 6.096 1.0160 5.125 0.00661 **
Residuals 13 2.577 0.1982
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
查看组间差异结果并作字母标注
library(agricolae)#需要用的包
data <- LSD.test(aov1,'trt',p.adj = 'bonferroni')#'bonferroni'#对P值进行修正
data
> data#注意这个里面内容比较多,比较杂,可以通过print(data$groups)命令只查看组间差异结果
$statistics
MSerror Df Mean CV
0.1982471 13 2.867345 15.52828
$parameters
test p.ajusted name.t ntr alpha
Fisher-LSD bonferroni trt 7 0.05
$means
val std r se LCL UCL Min Max Q25 Q50 Q75
a 3.062863 0.12408098 2 0.3148389 2.382695 3.743032 2.975125 3.150602 3.018994 3.062863 3.106733
b 3.542761 0.66056761 3 0.2570649 2.987406 4.098116 3.068194 4.297190 3.165547 3.262900 3.780045
c 2.625848 0.29751332 3 0.2570649 2.070493 3.181203 2.397264 2.962235 2.457654 2.518045 2.740140
d 2.234612 0.06537102 3 0.2570649 1.679257 2.789967 2.169607 2.300343 2.201747 2.233887 2.267115
e 2.139092 0.33342770 3 0.2570649 1.583737 2.694447 1.806937 2.473778 1.971749 2.136561 2.305169
f 3.614912 0.79146850 3 0.2570649 3.059557 4.170267 2.711331 4.185355 3.329690 3.948050 4.066702
k 2.916497 0.11905604 3 0.2570649 2.361142 3.471852 2.785514 3.018140 2.865676 2.945837 2.981988
$comparison
NULL
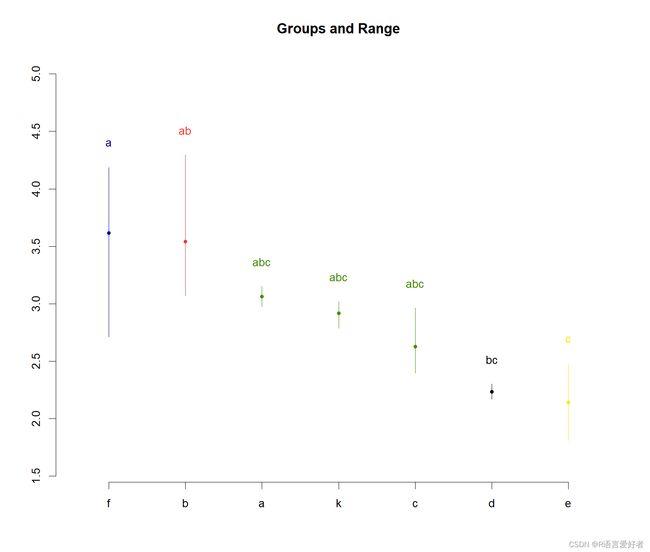
$groups
val groups
f 3.614912 a
b 3.542761 ab
a 3.062863 abc
k 2.916497 abc
c 2.625848 abc
d 2.234612 bc
e 2.139092 c
attr(,"class")
[1] "group"
#下面这个是单独查看groups的内容
#下面这个是单独查看groups的内容
print(data$groups)
> print(data$groups)
val groups
f 3.614912 a
b 3.542761 ab
a 3.062863 abc
k 2.916497 abc
c 2.625848 abc
d 2.234612 bc
e 2.139092 c
接下来对上述结果进行详细解释:
groups这一列的结果可以理解为找同类,其中val这列是按照均值从大到小排列,先把最大的标记为a,然后,找f的同类,凡是同类都标为a,直到找到第一个异类,然后标记为b,同时停止往下再找f的同类了,然后,开始找标记为b的同类,也就是d: 2.234612同类,先往上找同类,找到的都标为b,直到找完为止,然后再往下找同类,直到找到第一个异类,然后标记为c,然后重复这种工作。最后,这个同类就是两者间是不否存在差异显著性,异类就是存在差异显著性。
最后,画图
plot(data)
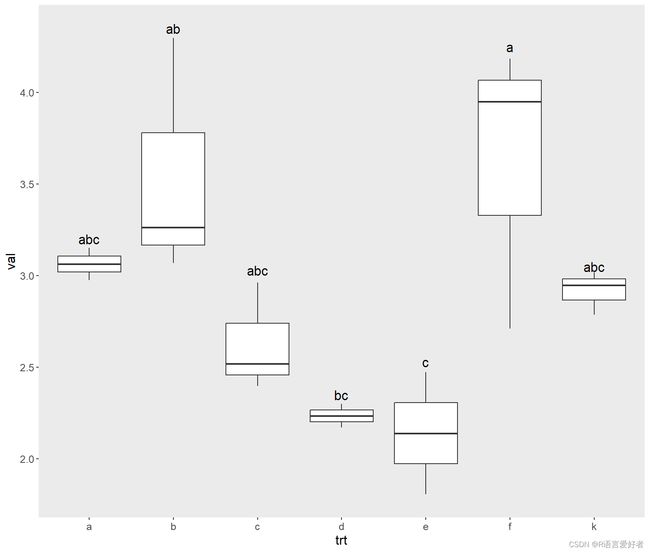
p3=ggplot(df,aes(x=trt,y=val))+ geom_boxplot()+
theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
text=element_text(size=13.5),
legend.position="None",
legend.title= element_blank(),)+
labs(y='val',x='trt')+
annotate("text", label = "abc",x = 1, y = 3.2, size = 5)+
annotate("text", label = "ab",x = 2, y = 4.35, size = 5)+
annotate("text", label = "abc",x = 3, y = 3.03, size = 5)+
annotate("text", label = "bc",x = 4, y = 2.35, size = 5)+
annotate("text", label = "c",x = 5, y = 2.53, size = 5)+
annotate("text", label = "a",x = 6, y = 4.25, size = 5)+
annotate("text", label = "abc",x = 7, y = 3.05, size = 5)
p3