Python&aconda系列:conda踩坑总结(deeptools 篇)
conda踩坑总结(deeptools 篇)
- 一. conda踩坑总结
-
-
- 第一步,换回官方源
- 第二步,更新base环境
- 第三步,灵活设置优先级
- 第四步,确定要装的软件和哪个版本的python适配,创建环境
- 第五步,激活环境并进入,调取帮助文档查看软件是否安装成功
-
- 二. conda安装软件与环境管理
-
-
- 利用bioconda 安装生物信息软件
- 安装指定版本的软件
- 使用conda建立环境
-
-
- 处理环境,失败 ;关于 Solving environment: failed with initial frozen solve,有提供的资料(详情见 `五. Deeptools error - Solving environment: failed with initial frozen solve`)
- UnsatisfiableError:与已有环境中的python不兼容
-
-
- 三. Python包中__init__.py作用
-
-
- Pycharm下的package树结构:
- 在Finder中的目录结构:
-
- Python中的包和模块有两种导入方式:精确导入和模糊导入:
-
- 精确导入:
- 模糊导入:
- 总结:
-
- 四. Python中的包ImportError
-
-
- 前言
- Python中的包
- 实例
-
- 下面我们来看一下包导入的各种情况:
-
- 导入python自带包或外部包
- 导入本工程中的包
- 导入父包中的模块
- 导入子包中的模块
- 导入兄弟包中的模块
- 总结
-
- 五. Deeptools error - Solving environment: failed with initial frozen solve
- 六. Installing multiqc on Conda produces "UnsatisfiableError:"
-
-
- 问题:
- 回答
-
接
Python&aconda系列:conda踩坑记录1.An unexpected error has occurred. Conda has prepared the above report
Python&aconda系列:conda踩坑记录2.UnsatisfiableError: The following specifications were found to be incompa
一. conda踩坑总结
深呼吸,捋一遍
参考 二. conda安装软件与环境管理
二. conda安装软件与环境管理 这篇分享帮大忙了,谢谢原作者
第一步,换回官方源
(base) vip9t07@tpm9-desktop:~$ vim ~/.condarc
(base) vip9t07@tpm9-desktop:~$ conda config --show channels
channels:
- https://repo.anaconda.com/pkgs/free/linux-64/
- https://repo.anaconda.com/pkgs/main/linux-64/
- defaults
- https://mirrors.ustc.edu.cn/anaconda/pkgs/free/
- https://mirrors.ustc.edu.cn/anaconda/cloud/bioconda/
- https://mirrors.bfsu.edu.cn/anaconda/pkgs/main
- https://mirrors.bfsu.edu.cn/anaconda/pkgs/free
第二步,更新base环境
(base) vip9t07@tpm9-desktop:~$ conda update -n base conda
Collecting package metadata (current_repodata.json): done
Solving environment: done
## Package Plan ##
environment location: /home/data/vip9t07/miniconda3
added / updated specs:
- conda
The following packages will be downloaded:
package | build
---------------------------|-----------------
asn1crypto-0.22.0 | py36_0 148 KB https://repo.anaconda.com/pkgs/free
certifi-2021.5.30 | py36h06a4308_0 139 KB defaults
cffi-1.10.0 | py36_0 341 KB https://repo.anaconda.com/pkgs/free
chardet-3.0.4 | py36_0 190 KB https://repo.anaconda.com/pkgs/free
conda-4.10.1 | py36h06a4308_1 2.9 MB defaults
conda-package-handling-1.7.3| py36h27cfd23_1 882 KB defaults
cryptography-1.8.1 | py36_0 846 KB https://repo.anaconda.com/pkgs/free
idna-2.6 | py36_0 122 KB https://repo.anaconda.com/pkgs/free
libffi-3.2.1 | 1 38 KB https://repo.anaconda.com/pkgs/free
openssl-1.0.2l | 0 3.2 MB https://repo.anaconda.com/pkgs/free
packaging-16.8 | py36_0 30 KB https://repo.anaconda.com/pkgs/free
pip-9.0.1 | py36_1 1.7 MB https://repo.anaconda.com/pkgs/free
pycparser-2.18 | py36_0 167 KB https://repo.anaconda.com/pkgs/free
pyopenssl-17.0.0 | py36_0 76 KB https://repo.anaconda.com/pkgs/free
pyparsing-2.2.0 | py36_0 96 KB https://repo.anaconda.com/pkgs/free
pysocks-1.6.6 | py36_0 20 KB https://repo.anaconda.com/pkgs/free
readline-6.2 | 2 606 KB https://repo.anaconda.com/pkgs/free
requests-2.25.1 | pyhd3eb1b0_0 52 KB defaults
ruamel_yaml-0.11.14 | py36_1 401 KB https://repo.anaconda.com/pkgs/free
setuptools-36.4.0 | py36_1 563 KB https://repo.anaconda.com/pkgs/free
six-1.10.0 | py36_0 19 KB https://repo.anaconda.com/pkgs/free
sqlite-3.13.0 | 0 4.0 MB https://repo.anaconda.com/pkgs/free
tk-8.5.18 | 0 1.9 MB https://repo.anaconda.com/pkgs/free
tqdm-4.15.0 | py36_0 48 KB https://repo.anaconda.com/pkgs/free
urllib3-1.21.1 | py36_0 153 KB https://repo.anaconda.com/pkgs/free
wheel-0.29.0 | py36_0 88 KB https://repo.anaconda.com/pkgs/free
xz-5.2.3 | 0 667 KB https://repo.anaconda.com/pkgs/free
yaml-0.1.6 | 0 246 KB https://repo.anaconda.com/pkgs/free
zlib-1.2.11 | 0 109 KB https://repo.anaconda.com/pkgs/free
------------------------------------------------------------
Total: 19.5 MB
The following packages will be UPDATED:
certifi anaconda/cloud/conda-forge::certifi-2~ --> pkgs/main::certifi-2021.5.30-py36h06a4308_0
conda anaconda/cloud/conda-forge::conda-4.1~ --> pkgs/main::conda-4.10.1-py36h06a4308_1
conda-package-han~ anaconda/cloud/conda-forge::conda-pac~ --> pkgs/main::conda-package-handling-1.7.3-py36h27cfd23_1
The following packages will be SUPERSEDED by a higher-priority channel:
asn1crypto anaconda/pkgs/free --> pkgs/free
cffi anaconda/pkgs/free --> pkgs/free
chardet anaconda/pkgs/free --> pkgs/free
cryptography anaconda/pkgs/free --> pkgs/free
idna anaconda/pkgs/free --> pkgs/free
libffi anaconda/pkgs/free --> pkgs/free
openssl anaconda/pkgs/free --> pkgs/free
packaging anaconda/pkgs/free --> pkgs/free
pip anaconda/pkgs/free --> pkgs/free
pycparser anaconda/pkgs/free --> pkgs/free
pyopenssl anaconda/pkgs/free --> pkgs/free
pyparsing anaconda/pkgs/free --> pkgs/free
pysocks anaconda/pkgs/free --> pkgs/free
readline anaconda/pkgs/free --> pkgs/free
requests anaconda/cloud/conda-forge::requests-~ --> pkgs/main::requests-2.25.1-pyhd3eb1b0_0
ruamel_yaml anaconda/pkgs/free --> pkgs/free
setuptools anaconda/pkgs/free --> pkgs/free
six anaconda/pkgs/free --> pkgs/free
sqlite anaconda/pkgs/free --> pkgs/free
tk anaconda/pkgs/free --> pkgs/free
tqdm anaconda/pkgs/free --> pkgs/free
urllib3 anaconda/pkgs/free --> pkgs/free
wheel anaconda/pkgs/free --> pkgs/free
xz anaconda/pkgs/free --> pkgs/free
yaml anaconda/pkgs/free --> pkgs/free
zlib anaconda/pkgs/free --> pkgs/free
Proceed ([y]/n)? y
Downloading and Extracting Packages
cffi-1.10.0 | 341 KB | ################################################################################################################# | 100%
cryptography-1.8.1 | 846 KB | ################################################################################################################# | 100%
xz-5.2.3 | 667 KB | ################################################################################################################# | 100%
pysocks-1.6.6 | 20 KB | ################################################################################################################# | 100%
packaging-16.8 | 30 KB | ################################################################################################################# | 100%
pyparsing-2.2.0 | 96 KB | ################################################################################################################# | 100%
pyopenssl-17.0.0 | 76 KB | ################################################################################################################# | 100%
sqlite-3.13.0 | 4.0 MB | ################################################################################################################# | 100%
yaml-0.1.6 | 246 KB | ################################################################################################################# | 100%
ruamel_yaml-0.11.14 | 401 KB | ################################################################################################################# | 100%
readline-6.2 | 606 KB | ################################################################################################################# | 100%
conda-package-handli | 882 KB | ################################################################################################################# | 100%
asn1crypto-0.22.0 | 148 KB | ################################################################################################################# | 100%
pycparser-2.18 | 167 KB | ################################################################################################################# | 100%
wheel-0.29.0 | 88 KB | ################################################################################################################# | 100%
requests-2.25.1 | 52 KB | ################################################################################################################# | 100%
tk-8.5.18 | 1.9 MB | ################################################################################################################# | 100%
chardet-3.0.4 | 190 KB | ################################################################################################################# | 100%
certifi-2021.5.30 | 139 KB | ################################################################################################################# | 100%
conda-4.10.1 | 2.9 MB | ################################################################################################################# | 100%
tqdm-4.15.0 | 48 KB | ################################################################################################################# | 100%
idna-2.6 | 122 KB | ################################################################################################################# | 100%
urllib3-1.21.1 | 153 KB | ################################################################################################################# | 100%
openssl-1.0.2l | 3.2 MB | ################################################################################################################# | 100%
six-1.10.0 | 19 KB | ################################################################################################################# | 100%
zlib-1.2.11 | 109 KB | ################################################################################################################# | 100%
pip-9.0.1 | 1.7 MB | ################################################################################################################# | 100%
libffi-3.2.1 | 38 KB | ################################################################################################################# | 100%
setuptools-36.4.0 | 563 KB | ################################################################################################################# | 100%
Preparing transaction: done
Verifying transaction: done
Executing transaction: done
第三步,灵活设置优先级
(base) vip9t07@tpm9-desktop:~$ conda config --set channel_priority flexible
第四步,确定要装的软件和哪个版本的python适配,创建环境
(base) vip9t07@tpm9-desktop:~$ conda create -n py38 deeptools=3.5.0 python=3
Collecting package metadata (current_repodata.json): done
Solving environment: failed with repodata from current_repodata.json, will retry with next repodata source.
Collecting package metadata (repodata.json): done
Solving environment: done
## Package Plan ##
environment location: /home/data/vip9t07/miniconda3/envs/py38
added / updated specs:
- deeptools=3.5.0
- python=3
The following packages will be downloaded:
package | build
---------------------------|-----------------
blas-1.0 | mkl 6 KB defaults
bzip2-1.0.8 | h7b6447c_0 78 KB defaults
certifi-2021.5.30 | py37h06a4308_0 139 KB defaults
curl-7.71.1 | hbc83047_1 140 KB defaults
cycler-0.10.0 | py37_0 13 KB defaults
deeptools-3.5.0 | py_0 143 KB https://mirrors.ustc.edu.cn/anaconda/cloud/bioconda
deeptoolsintervals-0.1.9 | py37h516909a_0 72 KB https://mirrors.ustc.edu.cn/anaconda/cloud/bioconda
freetype-2.10.4 | h5ab3b9f_0 596 KB defaults
intel-openmp-2021.2.0 | h06a4308_610 1.3 MB defaults
jpeg-9b | h024ee3a_2 214 KB defaults
kiwisolver-1.3.1 | py37h2531618_0 80 KB defaults
krb5-1.18.2 | h173b8e3_0 1.3 MB defaults
lcms2-2.12 | h3be6417_0 312 KB defaults
libcurl-7.71.1 | h20c2e04_1 305 KB defaults
libedit-3.1.20210216 | h27cfd23_1 167 KB defaults
libgfortran-ng-7.3.0 | hdf63c60_0 1006 KB defaults
libpng-1.6.37 | hbc83047_0 278 KB defaults
libssh2-1.9.0 | h1ba5d50_1 269 KB defaults
libtiff-4.2.0 | h85742a9_0 502 KB defaults
libwebp-base-1.2.0 | h27cfd23_0 437 KB defaults
lz4-c-1.9.3 | h2531618_0 186 KB defaults
matplotlib-base-3.3.4 | py37h62a2d02_0 5.1 MB defaults
mkl-2021.2.0 | h06a4308_296 144.3 MB defaults
mkl-service-2.3.0 | py37h27cfd23_1 55 KB defaults
mkl_fft-1.3.0 | py37h42c9631_2 170 KB defaults
mkl_random-1.2.1 | py37ha9443f7_2 287 KB defaults
numpy-1.20.2 | py37h2d18471_0 23 KB defaults
numpy-base-1.20.2 | py37hfae3a4d_0 4.5 MB defaults
olefile-0.46 | py37_0 50 KB defaults
pillow-8.2.0 | py37he98fc37_0 622 KB defaults
pip-21.1.1 | py37h06a4308_0 1.8 MB defaults
plotly-4.14.3 | pyhd3eb1b0_0 3.6 MB defaults
py2bit-0.3.0 | py37h14c3975_2 22 KB https://mirrors.ustc.edu.cn/anaconda/cloud/bioconda
pybigwig-0.3.17 | py37hc013797_0 77 KB https://mirrors.ustc.edu.cn/anaconda/cloud/bioconda
pyparsing-2.4.7 | pyhd3eb1b0_0 59 KB defaults
pysam-0.15.3 | py37hda2845c_1 2.5 MB https://mirrors.ustc.edu.cn/anaconda/cloud/bioconda
python-3.7.10 | hdb3f193_0 45.2 MB defaults
python-dateutil-2.8.1 | pyhd3eb1b0_0 221 KB defaults
retrying-1.3.3 | py37_2 16 KB defaults
scipy-1.6.2 | py37had2a1c9_1 15.5 MB defaults
setuptools-52.0.0 | py37h06a4308_0 710 KB defaults
six-1.15.0 | py37h06a4308_0 27 KB defaults
tornado-6.1 | py37h27cfd23_0 589 KB defaults
zstd-1.4.9 | haebb681_0 480 KB defaults
------------------------------------------------------------
Total: 233.2 MB
The following NEW packages will be INSTALLED:
_libgcc_mutex pkgs/main/linux-64::_libgcc_mutex-0.1-main
blas pkgs/main/linux-64::blas-1.0-mkl
bzip2 pkgs/main/linux-64::bzip2-1.0.8-h7b6447c_0
ca-certificates pkgs/main/linux-64::ca-certificates-2021.5.25-h06a4308_1
certifi pkgs/main/linux-64::certifi-2021.5.30-py37h06a4308_0
curl pkgs/main/linux-64::curl-7.71.1-hbc83047_1
cycler pkgs/main/linux-64::cycler-0.10.0-py37_0
deeptools anaconda/cloud/bioconda/noarch::deeptools-3.5.0-py_0
deeptoolsintervals anaconda/cloud/bioconda/linux-64::deeptoolsintervals-0.1.9-py37h516909a_0
freetype pkgs/main/linux-64::freetype-2.10.4-h5ab3b9f_0
intel-openmp pkgs/main/linux-64::intel-openmp-2021.2.0-h06a4308_610
jpeg pkgs/main/linux-64::jpeg-9b-h024ee3a_2
kiwisolver pkgs/main/linux-64::kiwisolver-1.3.1-py37h2531618_0
krb5 pkgs/main/linux-64::krb5-1.18.2-h173b8e3_0
lcms2 pkgs/main/linux-64::lcms2-2.12-h3be6417_0
ld_impl_linux-64 pkgs/main/linux-64::ld_impl_linux-64-2.33.1-h53a641e_7
libcurl pkgs/main/linux-64::libcurl-7.71.1-h20c2e04_1
libdeflate anaconda/cloud/bioconda/linux-64::libdeflate-1.0-h14c3975_1
libedit pkgs/main/linux-64::libedit-3.1.20210216-h27cfd23_1
libffi pkgs/main/linux-64::libffi-3.3-he6710b0_2
libgcc-ng pkgs/main/linux-64::libgcc-ng-9.1.0-hdf63c60_0
libgfortran-ng pkgs/main/linux-64::libgfortran-ng-7.3.0-hdf63c60_0
libpng pkgs/main/linux-64::libpng-1.6.37-hbc83047_0
libssh2 pkgs/main/linux-64::libssh2-1.9.0-h1ba5d50_1
libstdcxx-ng pkgs/main/linux-64::libstdcxx-ng-9.1.0-hdf63c60_0
libtiff pkgs/main/linux-64::libtiff-4.2.0-h85742a9_0
libwebp-base pkgs/main/linux-64::libwebp-base-1.2.0-h27cfd23_0
lz4-c pkgs/main/linux-64::lz4-c-1.9.3-h2531618_0
matplotlib-base pkgs/main/linux-64::matplotlib-base-3.3.4-py37h62a2d02_0
mkl pkgs/main/linux-64::mkl-2021.2.0-h06a4308_296
mkl-service pkgs/main/linux-64::mkl-service-2.3.0-py37h27cfd23_1
mkl_fft pkgs/main/linux-64::mkl_fft-1.3.0-py37h42c9631_2
mkl_random pkgs/main/linux-64::mkl_random-1.2.1-py37ha9443f7_2
ncurses pkgs/main/linux-64::ncurses-6.2-he6710b0_1
numpy pkgs/main/linux-64::numpy-1.20.2-py37h2d18471_0
numpy-base pkgs/main/linux-64::numpy-base-1.20.2-py37hfae3a4d_0
olefile pkgs/main/linux-64::olefile-0.46-py37_0
openssl pkgs/main/linux-64::openssl-1.1.1k-h27cfd23_0
pillow pkgs/main/linux-64::pillow-8.2.0-py37he98fc37_0
pip pkgs/main/linux-64::pip-21.1.1-py37h06a4308_0
plotly pkgs/main/noarch::plotly-4.14.3-pyhd3eb1b0_0
py2bit anaconda/cloud/bioconda/linux-64::py2bit-0.3.0-py37h14c3975_2
pybigwig anaconda/cloud/bioconda/linux-64::pybigwig-0.3.17-py37hc013797_0
pyparsing pkgs/main/noarch::pyparsing-2.4.7-pyhd3eb1b0_0
pysam anaconda/cloud/bioconda/linux-64::pysam-0.15.3-py37hda2845c_1
python pkgs/main/linux-64::python-3.7.10-hdb3f193_0
python-dateutil pkgs/main/noarch::python-dateutil-2.8.1-pyhd3eb1b0_0
readline pkgs/main/linux-64::readline-8.1-h27cfd23_0
retrying pkgs/main/linux-64::retrying-1.3.3-py37_2
scipy pkgs/main/linux-64::scipy-1.6.2-py37had2a1c9_1
setuptools pkgs/main/linux-64::setuptools-52.0.0-py37h06a4308_0
six pkgs/main/linux-64::six-1.15.0-py37h06a4308_0
sqlite pkgs/main/linux-64::sqlite-3.35.4-hdfb4753_0
tk pkgs/main/linux-64::tk-8.6.10-hbc83047_0
tornado pkgs/main/linux-64::tornado-6.1-py37h27cfd23_0
wheel pkgs/main/noarch::wheel-0.36.2-pyhd3eb1b0_0
xz pkgs/main/linux-64::xz-5.2.5-h7b6447c_0
zlib pkgs/main/linux-64::zlib-1.2.11-h7b6447c_3
zstd pkgs/main/linux-64::zstd-1.4.9-haebb681_0
Proceed ([y]/n)? y
Downloading and Extracting Packages
scipy-1.6.2 | 15.5 MB | ################################################################################################################# | 100%
intel-openmp-2021.2. | 1.3 MB | ################################################################################################################# | 100%
mkl_fft-1.3.0 | 170 KB | ################################################################################################################# | 100%
tornado-6.1 | 589 KB | ################################################################################################################# | 100%
blas-1.0 | 6 KB | ################################################################################################################# | 100%
bzip2-1.0.8 | 78 KB | ################################################################################################################# | 100%
deeptoolsintervals-0 | 72 KB | ################################################################################################################# | 100%
mkl-service-2.3.0 | 55 KB | ################################################################################################################# | 100%
pysam-0.15.3 | 2.5 MB | ################################################################################################################# | 100%
mkl_random-1.2.1 | 287 KB | ################################################################################################################# | 100%
pip-21.1.1 | 1.8 MB | ################################################################################################################# | 100%
pillow-8.2.0 | 622 KB | ################################################################################################################# | 100%
libtiff-4.2.0 | 502 KB | ################################################################################################################# | 100%
curl-7.71.1 | 140 KB | ################################################################################################################# | 100%
libgfortran-ng-7.3.0 | 1006 KB | ################################################################################################################# | 100%
zstd-1.4.9 | 480 KB | ################################################################################################################# | 100%
mkl-2021.2.0 | 144.3 MB | ################################################################################################################# | 100%
krb5-1.18.2 | 1.3 MB | ################################################################################################################# | 100%
numpy-1.20.2 | 23 KB | ################################################################################################################# | 100%
deeptools-3.5.0 | 143 KB | ################################################################################################################# | 100%
jpeg-9b | 214 KB | ################################################################################################################# | 100%
certifi-2021.5.30 | 139 KB | ################################################################################################################# | 100%
setuptools-52.0.0 | 710 KB | ################################################################################################################# | 100%
libcurl-7.71.1 | 305 KB | ################################################################################################################# | 100%
python-dateutil-2.8. | 221 KB | ################################################################################################################# | 100%
olefile-0.46 | 50 KB | ################################################################################################################# | 100%
py2bit-0.3.0 | 22 KB | ################################################################################################################# | 100%
six-1.15.0 | 27 KB | ################################################################################################################# | 100%
python-3.7.10 | 45.2 MB | ################################################################################################################# | 100%
pybigwig-0.3.17 | 77 KB | ################################################################################################################# | 100%
matplotlib-base-3.3. | 5.1 MB | ################################################################################################################# | 100%
libwebp-base-1.2.0 | 437 KB | ################################################################################################################# | 100%
cycler-0.10.0 | 13 KB | ################################################################################################################# | 100%
kiwisolver-1.3.1 | 80 KB | ################################################################################################################# | 100%
plotly-4.14.3 | 3.6 MB | ################################################################################################################# | 100%
pyparsing-2.4.7 | 59 KB | ################################################################################################################# | 100%
freetype-2.10.4 | 596 KB | ################################################################################################################# | 100%
numpy-base-1.20.2 | 4.5 MB | ################################################################################################################# | 100%
libpng-1.6.37 | 278 KB | ################################################################################################################# | 100%
libedit-3.1.20210216 | 167 KB | ################################################################################################################# | 100%
lcms2-2.12 | 312 KB | ################################################################################################################# | 100%
libssh2-1.9.0 | 269 KB | ################################################################################################################# | 100%
retrying-1.3.3 | 16 KB | ################################################################################################################# | 100%
lz4-c-1.9.3 | 186 KB | ################################################################################################################# | 100%
Preparing transaction: done
Verifying transaction: done
Executing transaction: done
#
# To activate this environment, use
#
# $ conda activate py38
#
# To deactivate an active environment, use
#
# $ conda deactivate
第五步,激活环境并进入,调取帮助文档查看软件是否安装成功
(base) vip9t07@tpm9-desktop:~$ conda activate py38
(py38) vip9t07@tpm9-desktop:~$ conda list
# packages in environment at /home/data/vip9t07/miniconda3/envs/py38:
#
# Name Version Build Channel
_libgcc_mutex 0.1 main defaults
blas 1.0 mkl defaults
bzip2 1.0.8 h7b6447c_0 defaults
ca-certificates 2021.5.25 h06a4308_1 defaults
certifi 2021.5.30 py37h06a4308_0 defaults
curl 7.71.1 hbc83047_1 defaults
cycler 0.10.0 py37_0 defaults
deeptools 3.5.0 py_0 https://mirrors.ustc.edu.cn/anaconda/cloud/bioconda
deeptoolsintervals 0.1.9 py37h516909a_0 https://mirrors.ustc.edu.cn/anaconda/cloud/bioconda
freetype 2.10.4 h5ab3b9f_0 defaults
intel-openmp 2021.2.0 h06a4308_610 defaults
jpeg 9b h024ee3a_2 defaults
kiwisolver 1.3.1 py37h2531618_0 defaults
krb5 1.18.2 h173b8e3_0 defaults
lcms2 2.12 h3be6417_0 defaults
ld_impl_linux-64 2.33.1 h53a641e_7 defaults
libcurl 7.71.1 h20c2e04_1 defaults
libdeflate 1.0 h14c3975_1 https://mirrors.ustc.edu.cn/anaconda/cloud/bioconda
libedit 3.1.20210216 h27cfd23_1 defaults
libffi 3.3 he6710b0_2 defaults
libgcc-ng 9.1.0 hdf63c60_0 defaults
libgfortran-ng 7.3.0 hdf63c60_0 defaults
libpng 1.6.37 hbc83047_0 defaults
libssh2 1.9.0 h1ba5d50_1 defaults
libstdcxx-ng 9.1.0 hdf63c60_0 defaults
libtiff 4.2.0 h85742a9_0 defaults
libwebp-base 1.2.0 h27cfd23_0 defaults
lz4-c 1.9.3 h2531618_0 defaults
matplotlib-base 3.3.4 py37h62a2d02_0 defaults
mkl 2021.2.0 h06a4308_296 defaults
mkl-service 2.3.0 py37h27cfd23_1 defaults
mkl_fft 1.3.0 py37h42c9631_2 defaults
mkl_random 1.2.1 py37ha9443f7_2 defaults
ncurses 6.2 he6710b0_1 defaults
numpy 1.20.2 py37h2d18471_0 defaults
numpy-base 1.20.2 py37hfae3a4d_0 defaults
olefile 0.46 py37_0 defaults
openssl 1.1.1k h27cfd23_0 defaults
pillow 8.2.0 py37he98fc37_0 defaults
pip 21.1.1 py37h06a4308_0 defaults
plotly 4.14.3 pyhd3eb1b0_0 defaults
py2bit 0.3.0 py37h14c3975_2 https://mirrors.ustc.edu.cn/anaconda/cloud/bioconda
pybigwig 0.3.17 py37hc013797_0 https://mirrors.ustc.edu.cn/anaconda/cloud/bioconda
pyparsing 2.4.7 pyhd3eb1b0_0 defaults
pysam 0.15.3 py37hda2845c_1 https://mirrors.ustc.edu.cn/anaconda/cloud/bioconda
python 3.7.10 hdb3f193_0 defaults
python-dateutil 2.8.1 pyhd3eb1b0_0 defaults
readline 8.1 h27cfd23_0 defaults
retrying 1.3.3 py37_2 defaults
scipy 1.6.2 py37had2a1c9_1 defaults
setuptools 52.0.0 py37h06a4308_0 defaults
six 1.15.0 py37h06a4308_0 defaults
sqlite 3.35.4 hdfb4753_0 defaults
tk 8.6.10 hbc83047_0 defaults
tornado 6.1 py37h27cfd23_0 defaults
wheel 0.36.2 pyhd3eb1b0_0 defaults
xz 5.2.5 h7b6447c_0 defaults
zlib 1.2.11 h7b6447c_3 defaults
zstd 1.4.9 haebb681_0 defaults
(py38) vip9t07@tpm9-desktop:~$ deeptools
usage:
> deeptools
[-h] [--version]
deepTools is a suite of python tools particularly developed for the efficient analysis of
high-throughput sequencing data, such as ChIP-seq, RNA-seq or MNase-seq.
Each tool should be called by its own name as in the following example:
$ bamCoverage -b reads.bam -o coverage.bw
=分割线==
大E了,新的报错很快啊,没有闪
Collecting package metadata (current_repodata.json): done
Solving environment: failed with initial frozen solve. Retrying with flexible solve.
Solving environment: failed with repodata from current_repodata.json, will retry with next repodata source.
Collecting package metadata (repodata.json): done
Solving environment: failed with initial frozen solve. Retrying with flexible solve.
Solving environment: \
Found conflicts! Looking for incompatible packages.
This can take several minutes. Press CTRL-C to abort.
failed
照旧,开始查
参考
python - 六. Installing multiqc on Conda produces "UnsatisfiableError:"
Conda incompatibility with CUDA · Issue #1284 · ewels/MultiQC (github.com)
(py38) vip9t07@tpm9-desktop:~$ conda config --prepend channels bioconda --prepend channels conda-forge
(py38) vip9t07@tpm9-desktop:~$ conda config --show channels
channels:
- conda-forge
- bioconda
- defaults
- https://repo.anaconda.com/pkgs/main/linux-64/
- https://repo.anaconda.com/pkgs/free/linux-64/
- https://mirrors.ustc.edu.cn/anaconda/pkgs/free/
- https://mirrors.ustc.edu.cn/anaconda/cloud/bioconda/
- https://mirrors.bfsu.edu.cn/anaconda/pkgs/main
- https://mirrors.bfsu.edu.cn/anaconda/pkgs/free
(py38) vip9t07@tpm9-desktop:~$ conda install multiqc
#中间略
Preparing transaction: done
Verifying transaction: done
Executing transaction: done
二. conda安装软件与环境管理
利用bioconda 安装生物信息软件
通过conda安装软件,首先从官网查找该软件是否被conda支持;若可以:
输入以下的命令进行安装:
conda install -y fastqc(软件名)
使用
which softname
或者
find -name softname
可以查看软件位置
安装指定版本的软件
conda install softname=1.0 1.0代指版本
先使用conda search softname参看当先版本。
使用conda建立环境
conda可以建立不同的环境,每种环境单独存在,互不干扰。
最好为不同的处理和软件建立不同的环境conda官网说明
建立一个新环境
conda create -n myenv #参数-n代表设置环境名称,myenv是具体的环境名,可以替换成自己想要的名称
建立一个新环境,同时指定该坏境中python的版本(看软件的要求)
conda create -n myenv python=2.7
还可以建立环境的同时安装软件
conda create -n myenv
conda create -n myenv Scipy=0.15.0
环境建立成功后,会提示环境的激活和关闭的方法 #要记住
source activate myenv
deactivate myenv
查看已有的环境
conda info --envs
删除某个环境
conda remove -n myenv --all
或者更加粗暴
rm -rf ~/miniconda3/envs/myenv/ #该路径是要删除的环境所在的路径
在这里我想分享一下之前自己遇到的一个BUG;# 与软件以及Python版本密切相关(虽然到现在我也不知道是为什么,但是希望我给出的方案能够帮到和我一样情况的小白)
-
非root
-
权限不足
-
报错 ImportError
-
Python中的包(详情见
三. Python包中__init__.py作用)
当时遇到这个问题,查了一些网上的资料,并没有解决;于是请教了一位老师,给出的点拨是:是服务器的依赖库有问题;导致的软件没发使用。
现在仔细的检查报错的信息,发现__init__.py的标识(详情见 四. Python中的包ImportError );补充一点知识点:init.py是Python中package的标识。
报错情况:
(chipseq) [yangjy@GSCG01 ~]$ bamCoverage -h
Traceback (most recent call last):
File "/home/yangjy/miniconda3/envs/chipseq/bin/bamCoverage", line 6, in <module>
from deeptools import writeBedGraph
File "/home/yangjy/miniconda3/envs/chipseq/lib/python2.7/site-packages/deeptools/writeBedGraph.py", line 8, in <module>
from deeptools.countReadsPerBin import getCoverageOfRegion, getSmoothRange
File "/home/yangjy/miniconda3/envs/chipseq/lib/python2.7/site-packages/deeptools/countReadsPerBin.py", line 7, in <module>
import bamHandler
File "/home/yangjy/miniconda3/envs/chipseq/lib/python2.7/site-packages/deeptools/bamHandler.py", line 3, in <module>
import pysam
File "/home/yangjy/miniconda3/envs/chipseq/lib/python2.7/site-packages/pysam/__init__.py", line 5, in <module>
from pysam.libchtslib import *
ImportError: libhts.so.2: cannot open shared object file: No such file or directory
正常情况:
(deeptools) [yangjy@GSCG01 ~]$ bamCoverage -h
usage: An example usage is:$ bamCoverage -b reads.bam -o coverage.bw
This tool takes an alignment of reads or fragments as input (BAM file) and generates a coverage track (bigWig or bedGraph) as output. The coverage is
calculated as the number of reads per bin, where bins are short consecutive counting windows of a defined size. It is possible to extended the length of
the reads to better reflect the actual fragment length. *bamCoverage* offers normalization by scaling factor, Reads Per Kilobase per Million mapped reads
(RPKM), counts per million (CPM), bins per million mapped reads (BPM) and 1x depth (reads per genome coverage, RPGC).
Required arguments:
--bam BAM file, -b BAM file
BAM file to process (default: None)
Output:
--outFileName FILENAME, -o FILENAME
Output file name. (default: None)
--outFileFormat {bigwig,bedgraph}, -of {bigwig,bedgraph}
Output file type. Either "bigwig" or "bedgraph". (default: bigwig)
Optional arguments:
--help, -h show this help message and exit
--scaleFactor SCALEFACTOR
The computed scaling factor (or 1, if not applicable) will be multiplied by this. (Default: 1.0)
Read coverage normalization options:
--effectiveGenomeSize EFFECTIVEGENOMESIZE
The effective genome size is the portion of the genome that is mappable. Large fractions of the genome are stretches of NNNN that
should be discarded. Also, if repetitive regions were not included in the mapping of reads, the effective genome size needs to be
adjusted accordingly. A table of values is available here:
http://deeptools.readthedocs.io/en/latest/content/feature/effectiveGenomeSize.html . (default: None)
Read processing options:
--extendReads [INT bp], -e [INT bp]
This parameter allows the extension of reads to fragment size. If set, each read is extended, without exception. *NOTE*: This
feature is generally NOT recommended for spliced-read data, such as RNA-seq, as it would extend reads over skipped regions.
*Single-end*: Requires a user specified value for the final fragment length. Reads that already exceed this fragment length will
not be extended. *Paired-end*: Reads with mates are always extended to match the fragment size defined by the two read mates.
Unmated reads, mate reads that map too far apart (>4x fragment length) or even map to different chromosomes are treated like
single-end reads. The input of a fragment length value is optional. If no value is specified, it is estimated from the data (mean
of the fragment size of all mate reads). (default: False)
...........................
注:这里使用的是deeptools软件
可以注意到两个的conda环境不同;报错时我激活的是chipseq的conda环境;而正常时 我激活的是deeptools的conda环境----------这中间经历了什么呢?
首先,我使用conda重新安装了deeptools;
尝试刚刚的命令,同样的报错
老师让我查看了deeptools的版本
(chipseq) [yangjy@GSCG01 ~]$ conda list deeptools
# packages in environment at /home/yangjy/miniconda3/envs/chipseq:
#
# Name Version Build Channel
deeptools 1.5.9.1 0 https://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/bioconda
#重点来了:Version = 1.5.9.1
而最新的deeptools有:
(chipseq) [yangjy@GSCG01 ~]$ conda search deeptools
Loading channels: done
# Name Version Build Channel
deeptools 1.5.8.2 0 anaconda/cloud/bioconda
deeptools 1.5.9.1 0 anaconda/cloud/bioconda
deeptools 2.0.0 py27_0
.......................... 此处省略x.x.x版本 。。。en........
deeptools 3.4.2 py_0 anaconda/cloud/bioconda
deeptools 3.4.3 py_0 anaconda/cloud/bioconda
deeptools 3.5.0 py_0 anaconda/cloud/bioconda
所以我的版本是最低的;
于是我使用了
conda update deeptools
,but=====>
怎么也更新不了???
update deeptools
Preparing transaction: done
Verifying transaction: done
Executing transaction: done
(chipseq) [yangjy@GSCG01 ~]$ conda list deeptools
#
# Name Version Build Channel
deeptools 1.5.9.1 0 https://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/bioconda
再更新(重复了三次)还是一样的版本,还是不能bamCoverage -h正常的显示。
随后,使用
conda install deeptools=3.5.0#指定安装版本
but----it still Version = 1.5.9.1;
bug
(chipseq) [yangjy@GSCG01 ~]$ conda install deeptools=3.5.0
Solving environment: failed with initial frozen solve. Retrying with flexible solve.
Solving environment: failed with repodata from current_repodata.json, will retry with next repodata source.
Solving environment: failed with initial frozen solve. Retrying with flexible solve.
Solving environment: -
Found conflicts! Looking for incompatible packages. failed
UnsatisfiableError: The following specifications were found
to be incompatible with the existing python installation in your environment:
Specifications:
- deeptools=3.5.0 -> python[version='>=3']
Your python: python=2
If python is on the left-most side of the chain, that's the version you've asked for.
When python appears to the right, that indicates that the thing on the left is somehow
not available for the python version you are constrained to. Note that conda will not
change your python version to a different minor version unless you explicitly specify
that.
报错信息解读:
处理环境,失败 ;关于 Solving environment: failed with initial frozen solve,有提供的资料(详情见 五. Deeptools error - Solving environment: failed with initial frozen solve)
UnsatisfiableError:与已有环境中的python不兼容
细节就在这里:
-deeptools = 3.5.0-> python [version = '> = 3']
意思安装deeptools = 3.5.0需要python的版本大于3以上;而我现在chipseq环境中的python = 2
注意,除非明确指定,否则conda不会将python版本更改为其他版本。(也就是我建立的chipseq指定了python的版本)
了解了这些信息,于是,总结下来
(1)python的版本与软件版本的兼容性(管理Python)
(2)使用conda search package_name --info 来查看你想要安装的软件的配置细节。
(3)查看当前环境的Python版本:
(chipseq) [yangjy@GSCG01 ~]$ python --version #chipseq环境的Python版本
Python 2.7.15
(deeptools) [yangjy@GSCG01 ~]$ python --version #deeptools环境的Python版本
Python 3.8.6
所以,最后怎么处理上面的bug呢?
创建一个新的conda环境并安装
deeptoolsconda create -n deeptools deeptools=3.5.0 python=3.8.6
激活这个环境
conda activate deeptools
尝试
bamCoverage -h
参考网站:
biostars
三. Python包中__init__.py作用
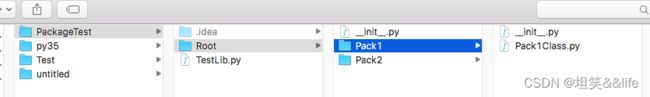
在创建python包的过程中,IDE都会在包根目录下创建一个__init__.py文件,该Python文件默认是空的.目录结构如下:
Pycharm下的package树结构:
在Finder中的目录结构:
从Finder中的目录就可以看出来,每个package实际上是一个目录(Directory),那么IDE是怎么识别它为package呢?
没错,__init__.py的第一个作用就是package的标识,如果没有该文件,该目录就不会认为是package。
Python中的包和模块有两种导入方式:精确导入和模糊导入:
精确导入:
from Root.Pack1 import Pack1Class
import Root.Pack1.Pack1Class
模糊导入:
from Root.Pack1 import *
模糊导入中的*中的模块是由__all__来定义的,__init__.py的另外一个作用就是定义package中的__all__,用来模糊导入,如__init__.py:
__all__ = ["Pack1Class","Pack1Class1"]
在包外部调用:
from Root.Pack1 import *
a = Pack1Class.Pack1_AA("Alvin")
a.PrintName()
init.py首先是一个python文件,所有还可以用来写python模块,但是不建议这么写,尽量保证__init__.py足够轻:
init.py:
__all__ = ["Pack1Class","Pack1Class1","Init_AA"]
class Init_AA:
def __init__(self,name):
self.name = name
def Greeting(self):
print("Hello ",self.name)
在测试中调用:
from Root.Pack1 import *
b = Init_AA("test")
b.Greeting()
总结:
从上边的例子可以看出,init.py的主要作用是:
-
Python中package的标识,不能删除
-
定义__all__用来模糊导入
-
编写Python代码(不建议在__init__中写python模块,可以在包中在创建另外的模块来写,尽量保证__init__.py简单)
四. Python中的包ImportError
前言
Python中的包给我提供了很好的代码组织,相似的功能模块放在同一个包内,不仅代码结构清晰,而且调用起来也比较方便(可以用*导入)
但是,我们在刚开始使用Python包的时候总是会遇到导入错误"ImportError: No module named ‘xxx’“,下面我们来简单介绍一下如何解决这类问题
Python中的包
Python中的包就和C#中的dll一样都是一些模块(或者类库)的集合。Python中新创建的包其实只有两个文件,一个是以包名命名的文件夹和其中的一个__init__.py文件(包的标识,可以到我之前的文章查看该文件的具体作用 三. Python包中__init__.py作用 )。既然了解了包的定义,那么我们如何引用包呢?在引用之前我们需要知道:
-
包是否为有效的包(用__init__.py标识)
-
包的路径在哪?是否添加到sys.path?
-
包中是否有要导入的模块
-
包的__init__.py中__all__是否包含要使用的模块(针对用from package import *导入的情况)
所以,当再次遇到导入错的时候,我们只需要检查以上四点,基本上能解决大部分的问题。
实例
下面我们来看一下包导入的各种情况:
导入python自带包或外部包
Python自带包可以直接用import package或者from package import module来进行导入,以为自带包都存在于系统包路径中,可以通过sys.path来查看:
>>> import sys
>>> sys.path
['', '/Users/xualvin/anaconda/lib/python35.zip', '/Users/xualvin/anaconda/lib/python3.5',
'/Users/xualvin/anaconda/lib/python3.5/plat-darwin', '/Users/xualvin/anaconda/lib/python3.5/lib-dynload',
'/Users/xualvin/anaconda/lib/python3.5/site-packages',
'/Users/xualvin/anaconda/lib/python3.5/site-packages/Sphinx-1.3.5-py3.5.egg',
'/Users/xualvin/anaconda/lib/python3.5/site-packages/aeosa',
'/Users/xualvin/anaconda/lib/python3.5/site-packages/setuptools-20.3-py3.5.egg']
第三方python包路径需要添加到sys.path或者复制到已有sys.path中才可以正常导入。使用sys.path.append(“path_to_third_party_packages”)进行添加
导入本工程中的包
如在上图包结构中的RootMain.py中导入Pack1,Pack1.SubPack和Pack2中的模块:
from Pack1 import *
from Pack1.SubPack1 import SubPack1_Module
from Pack2 import Pack2_Module
Pack1_Module.ModuleInfo()
SubPack1_Module.PrintInfo()
Pack2_Module.ModuleInfo()
其中Pack1中的__init__.py定义了__all__:
__all__ = ["SubPack1_Module"]
导入父包中的模块
导入父包中的模块,比如在Pack1.SubPack1中的SubPack1_Module.py调用Pack1中的Pack1_Module.py模块:
from Pack1 import Pack1_Module
def Pack1Module():
Pack1_Module.ModuleInfo()
导入子包中的模块
比如在Pack1_Module中导入SubPack1中的SubPack1_Module:
from Pack1.SubPack1 import SubPack1_Module
def InvokeSubPack1():
SubPack1_Module.PrintInfo()
导入兄弟包中的模块
这部分其实和第三方包的导入类似,我们需要将兄弟包导入到sys.path中,比如在Pack2中调用Pack1:
首先,在Pack2中导入Pack1的包路径,在Pack2中的__init__.py中导入(pycharm好像不需要这一步,VS2013的工程需要,不过加上没有什么错):
import sys
sys.path.append("/Users/xualvin/Bokeyuan/Pack1")
其次在Pack2中的模块中调用Pack1,导入方式和Python自带包一样:
from Pack1.SubPack1 import SubPack1_Module
from Pack1 import Pack1_Module
def InvokePack1():
Pack1_Module.ModuleInfo()
SubPack1_Module.PrintInfo()
总结
Python中的包导入我们只需要注意两点:一是导入的是不是包,二是导入的包路径是否能够被找到(sys.path或者本工程中),只要平时写程序的时候多注意,肯定可以避免很多麻烦。
五. Deeptools error - Solving environment: failed with initial frozen solve
Hi,
I am currently doing an ATAC-seq analysis for a bioinformatics course, but I am a python beginner and I am having trouble navigating it. I have 6 sorted bam files and my instructions say to use deeptools to assess the correlation between them (multiBamSummary -> plotCorrelation) and also check how much of the genome is covered and at what depth (plotCoverage).
I have been trying to install deeptools on Anaconda2 prompt (both on Python 3.7 and 2.7 versions) following this tutorial: https://deeptools.readthedocs.io/en/develop/content/installation.html First I had an environment error that I solved with the code below, but now I get other errors and I can not find a solution online? I tried uninstalling and reinstalling anaconda and other packages, but it does not work.
The code that I used:
conda create --name dianaenv python=2.7
conda activate dianaenv
conda config --add channels bioconda
conda install -c bioconda deeptools
The error: Collecting package metadata (current_repodata.json): done Solving environment: failed with initial frozen solve. Retrying with flexible solve. Solving environment: failed with repodata from current_repodata.json, will retry with next repodata source. Collecting package metadata (repodata.json): done Solving environment: failed with initial frozen solve. Retrying with flexible solve. Solving environment: / Found conflicts! Looking for incompatible packages. This can take several minutes. Press CTRL-C to abort.- failed
UnsatisfiableError: The following specifications were found to be incompatible with each other:
Package vs2008_runtime conflicts for: python=2.7 -> vs2008_runtime Package sqlite conflicts for: python=2.7 -> sqlite[version=‘>=3.25.3,<4.0a0|>=3.26.0,<4.0a0|>=3.27.2,<4.0a0|>=3.30.1,<4.0a0’] Package pysam conflicts for: deeptools -> pysam[version=‘>=0.14.0’] Package vc conflicts for: python=2.7 -> vc[version=‘9.*|>=9,<10.0a0’] Package deeptoolsintervals conflicts for: deeptools -> deeptoolsintervals[version=‘>=0.1.8’] Package matplotlib conflicts for: deeptools -> matplotlib[version=‘>=2.1.1|>=3.0.0’] Package pybigwig conflicts for: deeptools -> pybigwig[version=‘>=0.2.3’] Package ca-certificates conflicts for: python=2.7 -> ca-certificates Package scipy conflicts for: deeptools -> scipy[version=‘>=0.17.0’] Package py2bit conflicts for: deeptools -> py2bit[version=‘>=0.2.0’] Package plotly conflicts for: deeptools -> plotly[version=‘>=1.9.0|>=2.0.0’] Package pip conflicts for: python=2.7 -> pip Package numpy conflicts for: deeptools -> numpy[version=‘>=1.9.0’] Package pandas conflicts for: deeptools -> pandas
I also tried to install the requirements individually and installing numpy scipy and matplotlib works fine but when I try to install py2bit, pybigwig or pysam, I also get errors:
pip install py2bit DEPRECATION: Python 2.7 will reach the end of its life on January 1st, 2020. Please upgrade your Python as Python 2.7 won’t be maintained after that date. A future version of pip will drop support for Python 2.7. More details about Python 2 support in pip, can be found at https://pip.pypa.io/en/latest/development/release-process/#python-2-support Collecting py2bit Using cached https://files.pythonhosted.org/packages/53/bb/547a927bed736ead3dc909e1e552d57c9034bb9493eff80544c0cf6e4828/py2bit-0.3.0.tar.gz Building wheels for collected packages: py2bit Building wheel for py2bit setup.py) … error ERROR: Command errored out with exit status 1: command: ‘C:\Users\User\Anaconda2\envs\dianaenv\python.exe’ -u -c ‘import sys, setuptools, tokenize; sys.argv[0] = ‘"’“‘c:\users\user\appdata\local\temp\pip-install-6nxreu\py2bit\setup.py’”’“‘; file=’”‘“‘c:\users\user\appdata\local\temp\pip-install-6nxreu\py2bit\setup.py’”’“';f=getattr(tokenize, '”‘“‘open’”’“‘, open)(file);code=f.read().replace(’”‘"’\r\n’“'”‘, ‘"’"’\n’“'”‘);f.close();exec(compile(code, file, ‘"’“‘exec’”’"‘))’ bdist_wheel -d ‘c:\users\user\appdata\local\temp\pip-wheel-vrkfci’ --python-tag cp27 cwd: c:\users\user\appdata\local\temp\pip-install-6nxreu\py2bit\ Complete output (21 lines): C:\Users\User\Anaconda2\envs\dianaenv\lib\distutils\dist.py:267: UserWarning: Unknown distribution option: ‘classifier’ warnings.warn(msg) running bdist_wheel running build running build_py creating build creating build\lib.win-amd64-2.7 creating build\lib.win-amd64-2.7\py2bitTest copying py2bitTest\test.py -> build\lib.win-amd64-2.7\py2bitTest copying py2bitTest__init__.py -> build\lib.win-amd64-2.7\py2bitTest running egg_info writing py2bit.egg-info\PKG-INFO writing top-level names to py2bit.egg-info\top_level.txt writing dependency_links to py2bit.egg-info\dependency_links.txt reading manifest file ‘py2bit.egg-info\SOURCES.txt’ reading manifest template ‘MANIFEST.in’ writing manifest file ‘py2bit.egg-info\SOURCES.txt’ copying py2bitTest\foo.2bit -> build\lib.win-amd64-2.7\py2bitTest running build_ext building ‘py2bit’ extension error: Microsoft Visual C++ 9.0 is required. Get it from http://aka.ms/vcpython27
ERROR: Failed building wheel for py2bit Running setup.py clean for py2bit Failed to build py2bit Installing collected packages: py2bit Running setup.py install for py2bit … error ERROR: Command errored out with exit status 1: command: ‘C:\Users\User\Anaconda2\envs\dianaenv\python.exe’ -u -c ‘import sys, setuptools, tokenize; sys.argv[0] = ‘"’“‘c:\users\user\appdata\local\temp\pip-install-6nxreu\py2bit\setup.py’”’“‘; file=’”‘“‘c:\users\user\appdata\local\temp\pip-install-6nxreu\py2bit\setup.py’”’“';f=getattr(tokenize, '”‘“‘open’”’“‘, open)(file);code=f.read().replace(’”‘"’\r\n’“'”‘, ‘"’"’\n’“'”‘);f.close();exec(compile(code, file, ‘"’“‘exec’”’“‘))’ install --record ‘c:\users\user\appdata\local\temp\pip-record-xmdylh\install-record.txt’ --single-version-externally-managed --compile cwd: c:\users\user\appdata\local\temp\pip-install-6nxreu\py2bit\ Complete output (21 lines): C:\Users\User\Anaconda2\envs\dianaenv\lib\distutils\dist.py:267: UserWarning: Unknown distribution option: ‘classifier’ warnings.warn(msg) running install running build running build_py creating build creating build\lib.win-amd64-2.7 creating build\lib.win-amd64-2.7\py2bitTest copying py2bitTest\test.py -> build\lib.win-amd64-2.7\py2bitTest copying py2bitTest__init__.py -> build\lib.win-amd64-2.7\py2bitTest running egg_info writing py2bit.egg-info\PKG-INFO writing top-level names to py2bit.egg-info\top_level.txt writing dependency_links to py2bit.egg-info\dependency_links.txt reading manifest file ‘py2bit.egg-info\SOURCES.txt’ reading manifest template ‘MANIFEST.in’ writing manifest file ‘py2bit.egg-info\SOURCES.txt’ copying py2bitTest\foo.2bit -> build\lib.win-amd64-2.7\py2bitTest running build_ext building ‘py2bit’ extension error: Microsoft Visual C++ 9.0 is required. Get it from http://aka.ms/vcpython27 ---------------------------------------- ERROR: Command errored out with exit status 1: ‘C:\Users\User\Anaconda2\envs\dianaenv\python.exe’ -u -c 'import sys, setuptools, tokenize; sys.argv[0] = '”‘“‘c:\users\user\appdata\local\temp\pip-install-6nxreu\py2bit\setup.py’”’“‘; file=’”‘“‘c:\users\user\appdata\local\temp\pip-install-6nxreu\py2bit\setup.py’”’“';f=getattr(tokenize, '”‘“‘open’”’“‘, open)(file);code=f.read().replace(’”‘"’\r\n’“'”‘, ‘"’"’\n’“'”‘);f.close();exec(compile(code, file, ‘"’“‘exec’”’"‘))’ install --record ‘c:\users\user\appdata\local\temp\pip-record-xmdylh\install-record.txt’ --single-version-externally-managed --compile Check the logs for full command output.
Also, if I try to use pip install I get a matplotlib error: pip install --user deeptools DEPRECATION: Python 2.7 will reach the end of its life on January 1st, 2020. Please upgrade your Python as Python 2.7 won’t be maintained after that date. A future version of pip will drop support for Python 2.7. More details about Python 2 support in pip, can be found at https://pip.pypa.io/en/latest/development/release-process/#python-2-support Collecting deeptools Using cached https://files.pythonhosted.org/packages/82/8e/d9d4b66b2ce1bd48f1db43357c8eb019ae3e8bb1bb7a9e82667db981e1df/deepTools-3.3.1.tar.gz Collecting numpy>=1.9.0 Downloading https://files.pythonhosted.org/packages/48/83/203c397ecec78bdd618a0fb04a47482cfa2ae5ea2c6d428ed94258fe8671/numpy-1.16.5-cp27-cp27m-win_amd64.whl (11.9MB) |UUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUU| 11.9MB 2.5MB/s Collecting scipy>=0.17.0 Downloading https://files.pythonhosted.org/packages/49/73/22e125f335986fdc3b03c09cdf8ffe0d9d5471eec301aeb2e33db78b3e7f/scipy-1.2.2-cp27-cp27m-win_amd64.whl (30.5MB) |UUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUU| 30.5MB 187kB/s ERROR: Could not find a version that satisfies the requirement matplotlib>=3.0.0 (from deeptools) (from versions: 0.86, 0.86.1, 0.86.2, 0.91.0, 0.91.1, 1.0.1, 1.1.0, 1.1.1, 1.2.0, 1.2.1, 1.3.0, 1.3.1, 1.4.0, 1.4.1rc1, 1.4.1, 1.4.2, 1.4.3, 1.5.0, 1.5.1, 1.5.2, 1.5.3, 2.0.0b1, 2.0.0b2, 2.0.0b3, 2.0.0b4, 2.0.0rc1, 2.0.0rc2, 2.0.0, 2.0.1, 2.0.2, 2.1.0rc1, 2.1.0, 2.1.1, 2.1.2, 2.2.0rc1, 2.2.0, 2.2.2, 2.2.3, 2.2.4) ERROR: No matching distribution found for matplotlib>=3.0.0 (from deeptools)
-------------------------------------------------
Please highlight code, data examples and error messages with the code option (10101) to improve (or enable) readability.
---------------------------------------------------------------------------------------------------
First, you should make your error messages more clear by using strong or other marks to let us understand your question easier.
Second, Installing packages by pip in conda is strongly not recommended, see why here: using pip in a conda envrionment.
Since your problem is conda found conflict when sloving your environment, I think maybe you pip install something else previously, then conda didn’t know what you have done, which caused conflict.
I recommend you to completely uninstall your conda, then try all these things again.
---------------------------------------------------------------------------------------------------
六. Installing multiqc on Conda produces “UnsatisfiableError:”
问题:
I have tried installing a program called multiqc and it throws this error when I try to install it in my conda environment, I have tried to install alternative version of python contained in the list but it doesn’t appear to be working.
The command
conda install -c bioconda multiqc
The install
Collecting package metadata (current_repodata.json): done
Solving environment: failed with initial frozen solve. Retrying with flexible solve.
Solving environment: failed with repodata from current_repodata.json, will retry with next repodata source.
Collecting package metadata (repodata.json): done
Solving environment: failed with initial frozen solve. Retrying with flexible solve.
Solving environment: \
Found conflicts! Looking for incompatible packages.
This can take several minutes. Press CTRL-C to abort.
failed \
And finally the error
UnsatisfiableError: The following specifications were found
to be incompatible with the existing python installation in your environment:
Specifications:
- multiqc -> python[version='2.7.*|3.5.*|3.6.*|>=2.7,<2.8.0a0|>=3.5,<3.6.0a0|>=3.6,<3.7.0a0|3.4.*']
Your python: python=3.7
If python is on the left-most side of the chain, that's the version you've asked for.
When python appears to the right, that indicates that the thing on the left is somehow
not available for the python version you are constrained to. Note that conda will not
change your python version to a different minor version unless you explicitly specify
that.
FlyingTeller:How did you try to install an alternative version of python? Your error suggests that you are using 3.7 which seems to be incompatible with the package
nekomatic:Always, always try creating a new environment: conda create -n mynewenv -c bioconda multiqc and add any other packages you need on the same line after multiqc. conda will figure out what's compatible.
NajlaBioinfo:This command, (Ref. :multiqc.info/docs), conda install -c bioconda -c conda-forge multiqc works for me under "Debian GNU/Linux 10 (buster)" and "conda-4.8.3". Hope this helps. Good luck.
---------------------------------------------------------------------------------------------------
回答
The command to install MultiQC should not use -c bioconda. Instead, set up the conda channels as per the bioconda documentation and install without the -c flag:
# Only need to do this once
conda config --add channels defaults
conda config --add channels bioconda
conda config --add channels conda-forge
# Install MultiQC
conda install multiqc
If you prefer, you can also install from PyPI or multiple other sources: https://multiqc.info/docs/#installing-multiqc
MultiQC works and is tested on Python 3.6-3.9 at the time of writing. 3.7 should definitely work, so that’s not the issue.
---------------------------------------------------------------------------------------------------
小狼小狼_e211
conda踩坑总结
shine_9457
conda安装软件与环境管理添加链接描述
测试改进工场
Python包中__init__.py作用
Python中的包ImportError
biostars
Deeptools error - Solving environment: failed with initial frozen solve
stackoverflow
Installing multiqc on Conda produces “UnsatisfiableError:”