pie plot | ggplot2 画饼状图,排序并标注文字
难点,如何保证饼图的顺序?
难点2,如何保证饼图顺序和文字顺序一致?
解决方法:设置分类变量为因子。
1. 数据
table() |> as.data.frame() 获取统计变量,按Freq排序,对 Var1 和 anno 转为因子。
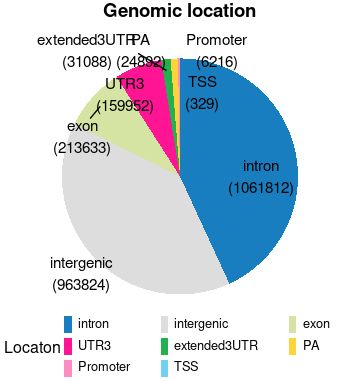
> dat
Freq Var1 anno
intron 1061812 intron intron\n(1061812)
intergenic 963824 intergenic intergenic\n(963824)
exon 213633 exon exon\n(213633)
UTR3 159952 UTR3 UTR3\n(159952)
extended3UTR 31088 extended3UTR extended3UTR\n(31088)
PA 24892 PA PA\n(24892)
Promoter 6216 Promoter Promoter\n(6216)
TSS 329 TSS TSS\n(329)
2.效果
3. 代码
# 去掉背景
library(ggplot2)
blank_theme = theme_minimal(base_size=12)+
theme(
axis.title.x = element_blank(),
axis.title.y = element_blank(),
axis.text.x = element_blank(),
axis.text.y = element_blank(),
panel.border = element_blank(),
panel.grid=element_blank(),
axis.ticks = element_blank(),
plot.title=element_text(size=14, face="bold",
hjust = 0.5)#大图标题居中
)
library(ggrepel) #geom_text_repel 比 geom_text新增功能:防止文字遮挡,自动连线点和标签
library(ggplot2)
ggPie=function(df2, colors=NULL, title="Genomic location"){
#df=df2
g=ggplot(df2, mapping=aes(x="", y=count,fill=type))+
geom_bar(stat="identity",width=0.5)+
#coord_polar("y", start=0)+ #0度角起始角度
coord_polar(theta = 'y', direction = -1)+ #direction设置方向
#scale_fill_manual(values=col6)
blank_theme+
theme(legend.position="bottom", #图例显示位置
legend.margin=margin(t = -1.5, unit='line'), #图例整体上边距,缩减n行
legend.spacing.x = unit(2, 'pt'), #图例之间的x距离
legend.spacing.y = unit(2,"pt"), #图例之间的y距离
#plot.margin=unit(c(1,0,1,0),"lines"), #图形外围边框top,right,bottom,left
legend.box = "horizontal", #多图排列方式 'horizontal', 'vertical'
#legend.direction="horizontal", #图例排列方向 'horizontal', 'vertical'
#legend.title.align = 0.5, #?
#legend.key = element_rect(color = NA, fill = NA), #图例每个方块的边框
#legend.key = element_rect(size = 1,color = 'yellow' ),
#legend.key.size = unit(0.2, "cm"), #图例方块大小
legend.key.height=unit(1,"line"), #图例方块的高度
legend.key.width=unit(0.5,"line"), #图例方块的宽度度
#标签right距离,left距离
legend.text = element_text(margin = margin(r = 20, l=2,t = -3, unit = "pt")),
#legend.text = element_text(margin = margin(t = -5,unit='pt')), #图例文字上移-n,或下移n
legend.background = element_blank() )+ #去掉图例背景
#guides(fill = guide_legend(title = NULL))+ #去掉图例标题
guides(fill = guide_legend(ncol = 3, #图例几列
inset=-0.5,
byrow=T))+ #图例横着排?默认竖着排
labs( #title="Pie plot (ggplot2)", #设置大图标题
#subtitle ="subtitles here, pie from count data.",
x="",y="")+
geom_text_repel(stat="identity",aes(x=1.2,y=count, label = tag), size=4, #饼图上的文字
position=position_stack(vjust = 0.5));g
#geom_text(stat="identity",aes(y=x, label = scales::percent(x/100)),
# size=4, position=position_stack(vjust = 0.5))
# set colors
if(is.null(colors)){
g=g+scale_fill_brewer("Locaton", palette="Set2")
}else{
g=g+scale_fill_manual("Locaton", values=colors)
}
return(g+ggtitle(title))
}
# input data
Input =("
Freq Var1
1061812 intron
963824 intergenic
213633 exon
159952 UTR3
31088 extended3UTR
24892 PA
6216 Promoter
329 TSS
")
dat=read.table(textConnection(Input),
header=TRUE,
row.names=2)
dat$Var1 = rownames(dat)
dat$anno = paste0(dat$Var1, "\n(", dat$Freq ,")")
# set as factor
dat$Var1 = factor(dat$Var1, levels = dat$Var1)
dat$anno = factor(dat$anno, levels = dat$anno)
dat
str(dat)
# default result
ggPie(data.frame(count=dat$Freq, type=dat$Var1, tag=dat$anno))
# set color for each location
color.dat=c("#FED439FF","#FF1493", "#22b14c", "#D5E4A2FF","#197EC0FF", "#71D0F5FF", "#FD8CC1", "#dddddd")
names(color.dat)=c("PA", "UTR3", "extended3UTR", "exon","intron", "TSS", "Promoter", "intergenic")
color.dat = color.dat[ rownames(dat) ]
color.dat
ggPie(data.frame(count=dat$Freq, type=dat$Var1, tag=dat$anno), colors=color.dat)
ggsave(filename ="/docker/jinwf/wangjl/scPolyA-seq2/chenxi/PBMC/annotate/pasPostions_GenomicLocation.pie.pdf", width=5.5, height=5)
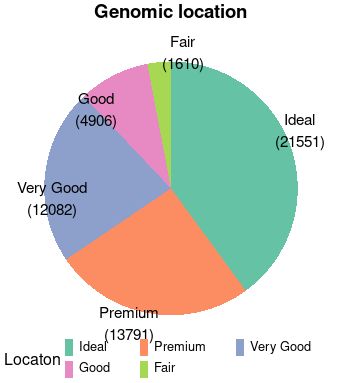
4. example2
#
dat=as.data.frame(table(diamonds$cut)); dat
dat$anno = paste0(dat$Var1, "\n(", dat$Freq ,")")
dat=dat[order(-dat$Freq),]
# to factor
dat$Var1 = factor(dat$Var1, levels = dat$Var1)
dat$anno = factor(dat$anno, levels = dat$anno)
dat
# plot
ggPie(data.frame(count=dat$Freq, type=dat$Var1, tag=dat$anno))