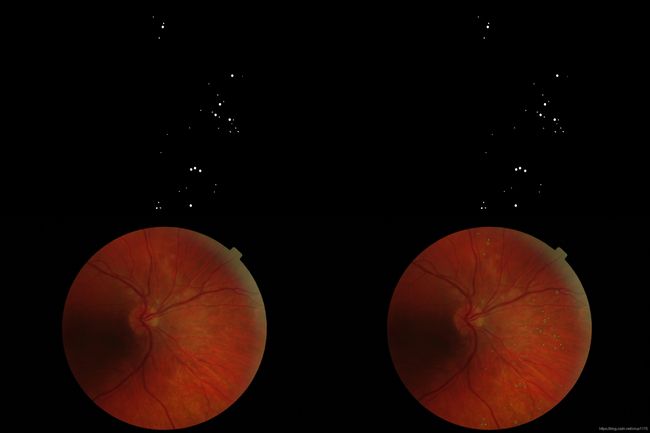
e_ophtha_MA眼底数据集—根据微血管瘤标注Mask绘制Contour轮廓图
眼底医学图像处理:微血管瘤Microaneurysm检测分割
采用数据集e_ophtha中的e_ophtha_MA,此数据集可从互联网下载
实现根据微血管瘤标注Mask,在原图绘制轮廓图,以直观了解微血管瘤,以便检测分割微血管瘤
1. 可展示数据集中原图和绘制轮廓图的并列拼接图
2. 可保存Mask,原图,根据标注绘制轮廓图的眼底图的拼接图
1. 原图和绘制轮廓图的并列拼接图
2. 保存Mask,原图,根据标注绘制轮廓图的眼底图的拼接图
3. 代码
# -*- coding: utf-8 -*-
'''
@Time : 2020.03.28 12:18
@Author : Shibing Xiang
@FileName: MaskinFundus.py
@Software: PyCharm
'''
import numpy as np
import cv2,os,imageio
from skimage import measure
def Union_Masks(rst, annotation):
print(type(rst))
def zh(string):
return string.encode("gbk").decode(errors="ignore")
def CVShow(img, title = None):
img = np.array(img)
if(len(img.shape) == 3):
h, w, d = img.shape
else:h, w = img.shape
fscale = 1800/w
cv2.namedWindow(title,0)
cv2.resizeWindow(title, np.int(w * fscale), np.int(h * fscale))
cv2.imshow(title, img)
key = cv2.waitKey(0)
if key == 27: # wait for ESC key to exit
cv2.destroyAllWindows()
return False
elif key == ord('s'):
cv2.destroyAllWindows()
return True
def read_directory(root_path,mask = False):
images = []
filepaths = []
names = os.listdir(root_path)
for filename in names:
if filename != 'Thumbs.db':
filepath = os.path.normpath(root_path + "/" + filename)
if mask:
img = cv2.imread(filepath, 0)
else:
img = cv2.imread(filepath)
filepaths.append(filepath)
images.append(img)
else: pass
return names, filepaths, np.array(images)
def CVdrawContour(mask,img, rect = False):
_, contours, hierarchy = cv2.findContours(mask, cv2.RETR_EXTERNAL, cv2.CHAIN_APPROX_SIMPLE)
if rect:
cv2.drawContours(img, contours, -1, (255, 0, 0), 1, hierarchy = hierarchy)
for cnt in contours:
x, y, w, h = cv2.boundingRect(cnt)
cv2.rectangle(img, (x, y), (x + w, y + h), (0, 255, 0), 1)
else: cv2.drawContours(img, contours, -1, (0, 255, 0), 1, hierarchy = hierarchy)
def skimContour(mask,img, color = (0, 176, 80)):
contours = measure.find_contours(mask, 0.5)
for c in contours:
c = np.around(c).astype(np.int)
img[c[:, 0], c[:, 1]] = np.array(color)
def MaskinFundus(masks, images, filenames, method = 'CV', show = False):
MaskedImages = images.copy() # 即使这样也会修改 images 的值
if method == 'CV':
for i in range(len(masks)):
CVdrawContour(masks[i], MaskedImages[i])
if show:
I_merge = np.hstack((images[i], MaskedImages[i])) # 水平拼接
CVShow(I_merge, 'The ' + str(i+1) + ' th - '+ filenames[i])
elif method == 'skimage':
for i in range(len(masks)):
skimContour(masks[i], MaskedImages[i])
if show:
I_merge = np.hstack((images[i], MaskedImages[i])) # 水平拼接
CVShow(I_merge, 'The ' + str(i + 1) + ' th - ' + filenames[i])
else: return None
return MaskedImages
if __name__ == '__main__':
root_path = r'J:\Image Projects\Microaneurysm Detection\Datasets\2013_e_ophtha\e_ophtha_MA'
for temp in os.listdir(root_path+'/'+'MA'):
Annotation_MA = os.path.normpath(root_path + '/Annotation_MA/' + temp)
MA = os.path.normpath(root_path + '/MA/' + temp)
_, _, masks = read_directory(Annotation_MA, mask=True)
filenames, _, images = read_directory(MA)
MaskedImages = MaskinFundus(masks, images, filenames, 'CV')
for i in range(len(masks)):
hmerge = np.hstack((images[i], MaskedImages[i])) # 水平拼接
save = CVShow(hmerge , temp + ' - ' + str(i + 1) + ' - ' + filenames[i])
if save:
hmerge0 = np.hstack((masks[i], masks[i])) # 水平拼接
hmerge0 = cv2.merge([hmerge0,hmerge0,hmerge0])
I_merge = np.vstack((hmerge0, hmerge)) # 垂直拼接
savepath = root_path + '/Sample Images/' + temp + '-' + filenames[i]
cv2.imwrite(savepath, I_merge)
# MA_masks = r'J:\Image Projects\Microaneurysm Detection\Datasets\2013_e_ophtha\e_ophtha_MA\Annotation_MA\E0001520'
# MA_imges = r'J:\Image Projects\Microaneurysm Detection\Datasets\2013_e_ophtha\e_ophtha_MA\MA\E0001520'
# _, masks = read_directory(MA_masks, mask=True)
# paths, images = read_directory(MA_imges)
# MaskinFundus(masks, images, 'CV')