windows下python3.4的scikit-learn搭建
环境:Windows64位(兼容32位的),Python3.4,Eclipse的PyDev。
前提:
Python的安装和Eclipse的PyDev安装请看:
Windows系统下Pythong环境的搭建:http://www.cnblogs.com/windinsky/archive/2012/09/20/2695520.html
Python+Eclipse+Pydev环境搭建:http://www.cnblogs.com/Bonker/p/3584707.html
我个人还碰到在Eclipse安装PyDev包的时候到一半就出现Error的情况,网上找资料应该是被墙的缘故吧,用以下方法解决了:
http://blog.csdn.net/alvine008/article/details/19749851
引用如下:
网上搜了一下,大概是由于pydev被和谐了,大概有两种解决思路
1,直接把pydev下载下来,直接手动安装。
到pydev的主页,选择自己python版本对应的dev版本,最好选一样版本号的,不然可能出现pydev选项出不来的错误。http://sourceforge.net/projects/pydev/files/pydev/
把下载好的pydev压缩包内的plugins和features 文件夹内的内容复制到eclipse的解压目录的对应的文件夹中即可。
2,给eclipse 设置代理
默认的Eclipse 是不用代理上网,但在一些公司的局域网,需要使用代理上网,
因而需要手工设置eclipse的上网设置
window-->preferences-->general-->network connections
选中 manual proxy configuration: 依次填入http proxy , port就ok 了。
另外,如果代理需要用帐号和密码就需要选中 Enable proxy authentication,
然后填上 user name 及password 取消,ok.
正题:
最近为了搞定项目中的蜜罐数据分析,需要机器学习和数据挖掘,然后找python的工具,看知乎有说scikit-learn不错,于是想搞搞
其官网为:http://scikit-learn.org,上面有很不错的学习资料,当然了,如果你英文好的话
比如,http://scikit-learn.org/stable/auto_examples/cluster/plot_kmeans_digits.html#example-cluster-plot-kmeans-digits-py
print(__doc__)
from time import time
import numpy as np
import matplotlib.pyplot as plt
from sklearn import metrics
from sklearn.cluster import KMeans
from sklearn.datasets import load_digits
from sklearn.decomposition import PCA
from sklearn.preprocessing import scale
np.random.seed(42)
digits = load_digits()
data = scale(digits.data)
n_samples, n_features = data.shape
n_digits = len(np.unique(digits.target))
labels = digits.target
sample_size = 300
print("n_digits: %d, \t n_samples %d, \t n_features %d"
% (n_digits, n_samples, n_features))
print(79 * '_')
print('% 9s' % 'init'
' time inertia homo compl v-meas ARI AMI silhouette')
def bench_k_means(estimator, name, data):
t0 = time()
estimator.fit(data)
print('% 9s %.2fs %i %.3f %.3f %.3f %.3f %.3f %.3f'
% (name, (time() - t0), estimator.inertia_,
metrics.homogeneity_score(labels, estimator.labels_),
metrics.completeness_score(labels, estimator.labels_),
metrics.v_measure_score(labels, estimator.labels_),
metrics.adjusted_rand_score(labels, estimator.labels_),
metrics.adjusted_mutual_info_score(labels, estimator.labels_),
metrics.silhouette_score(data, estimator.labels_,
metric='euclidean',
sample_size=sample_size)))
bench_k_means(KMeans(init='k-means++', n_clusters=n_digits, n_init=10),
name="k-means++", data=data)
bench_k_means(KMeans(init='random', n_clusters=n_digits, n_init=10),
name="random", data=data)
# in this case the seeding of the centers is deterministic, hence we run the
# kmeans algorithm only once with n_init=1
pca = PCA(n_components=n_digits).fit(data)
bench_k_means(KMeans(init=pca.components_, n_clusters=n_digits, n_init=1),
name="PCA-based",
data=data)
print(79 * '_')
###############################################################################
# Visualize the results on PCA-reduced data
reduced_data = PCA(n_components=2).fit_transform(data)
kmeans = KMeans(init='k-means++', n_clusters=n_digits, n_init=10)
kmeans.fit(reduced_data)
# Step size of the mesh. Decrease to increase the quality of the VQ.
h = .02 # point in the mesh [x_min, m_max]x[y_min, y_max].
# Plot the decision boundary. For that, we will assign a color to each
x_min, x_max = reduced_data[:, 0].min() + 1, reduced_data[:, 0].max() - 1
y_min, y_max = reduced_data[:, 1].min() + 1, reduced_data[:, 1].max() - 1
xx, yy = np.meshgrid(np.arange(x_min, x_max, h), np.arange(y_min, y_max, h))
# Obtain labels for each point in mesh. Use last trained model.
Z = kmeans.predict(np.c_[xx.ravel(), yy.ravel()])
# Put the result into a color plot
Z = Z.reshape(xx.shape)
plt.figure(1)
plt.clf()
plt.imshow(Z, interpolation='nearest',
extent=(xx.min(), xx.max(), yy.min(), yy.max()),
cmap=plt.cm.Paired,
aspect='auto', origin='lower')
plt.plot(reduced_data[:, 0], reduced_data[:, 1], 'k.', markersize=2)
# Plot the centroids as a white X
centroids = kmeans.cluster_centers_
plt.scatter(centroids[:, 0], centroids[:, 1],
marker='x', s=169, linewidths=3,
color='w', zorder=10)
plt.title('K-means clustering on the digits dataset (PCA-reduced data)\n'
'Centroids are marked with white cross')
plt.xlim(x_min, x_max)
plt.ylim(y_min, y_max)
plt.xticks(())
plt.yticks(())
plt.show()
这个KNN的例子中,把它代码放入PyDev中,好多import报错,于是安装开始
首先,scikit-learn需要前置的scipy和numpy,以及画图的matplotlib,而matplotlib又需要前置的six,deteutils,pyparsing,
所以总共需要安装7个。
参考资料:
http://scikit-learn.org/dev/install.html#install-by-distribution
http://www.mamicode.com/info-detail-511470.html
http://jingyan.baidu.com/article/454316ab4a5b61f7a7c03a96.html
http://blog.csdn.net/pfanaya/article/details/7451815
安装:
numpy(.exe格式)下载直接安装
http://sourceforge.net/projects/numpy/files/NumPy
scipy(.exe格式)下载直接安装
http://sourceforge.net/projects/scipy/files/
six(.tar.gz格式)先解压到A文件夹,然后在cmd下进入A文件夹输入A:\> C:\Python34\python setup.py install
https://pypi.python.org/pypi/six
deteutils(.tar.gz格式)先解压到A文件夹,然后在cmd下进入A文件夹输入A:\> C:\Python34\python setup.py install
https://pypi.python.org/pypi/dateutils
pyparsing(.exe格式)下载直接安装
https://pypi.python.org/pypi/pyparsing/2.0.3
matplotlib(.exe格式)下载直接安装
https://pypi.python.org/pypi/matplotlib
最终为了搞定
scikit-learn(.exe格式)下载直接安装,也可以直接用pip命令,cmd下进入python34安装目录下的Scripts的文件夹,然后
pip install -U scikit-learn
https://pypi.python.org/pypi/scikit-learn/0.16.1
其中,除了最后一个scikit-learn我是直接安装的,其他的安装包(.exe或.tar.gz)我都已经传到360云盘了,分享如下:
文件:[Apie]scikit-learn安装的东东.zip
http://yunpan.cn/cjEDJzUmFhkiT 访问密码 e26b
结果安装完了之后,还是有一个出错:
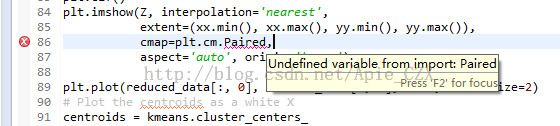
在第84行中出现报错:
plt.imshow(Z, interpolation='nearest',
extent=(xx.min(), xx.max(), yy.min(), yy.max()),
cmap=plt.cm.Paired,
aspect='auto', origin='lower')
报错信息:Undefined variable from import: Paired
查阅了资料可以搞定:
http://stackoverflow.com/questions/2112715/how-do-i-fix-pydev-undefined-variable-from-import-errors
I removed these errors altogether by going to:
Window -> Preferences -> PyDev -> Editor -> Code Analysis -> Undefined -> Undefined Variable From Import -> Ignore
And that's that.
It may also be, Window -> Preferences -> PyDev -> Editor -> Code Analysis -> Imports -> Import not found -> Ignore
(当然,后来发现了其实让它一直错着也照样可以运行,囧~~)
于是,就可以跑出来了。
P.S:
网上还有whl,easy_install(需要setuptools)之类的,有些是针对python2.7或其他操作系统平台的,我没有尝试过。
——Apie陈小旭
