漂亮,美观的图表之Matlab强势回归~~~~走你5
本文将学习三个图形函数——stem, staris,pie.
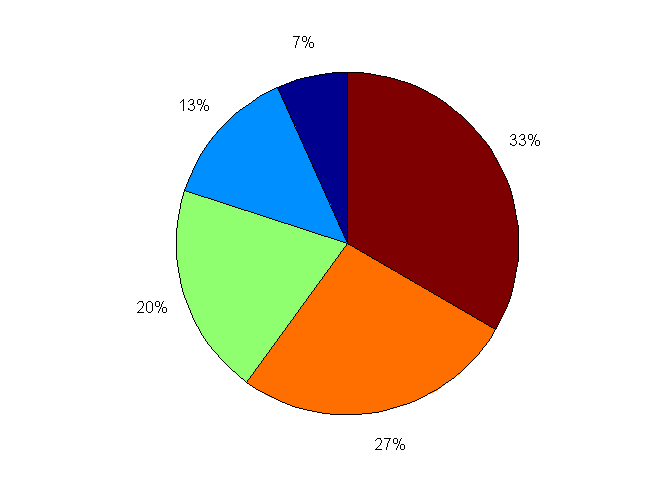
%% pie用于描绘平面饼图 X = [1, 2, 3, 4, 5]; pie(X)
%% 画饼状图
Expenses = [20 10 40 12 20 19 5 15];
ExpenseCategories = {'Food','Medical','Lodging','Incidentals','Transport','Utilities','Gifts','Shopping'};
MostLeastExpensive = (Expenses==max(Expenses)|Expenses==min(Expenses));
h=pie(gca,Expenses,MostLeastExpensive,ExpenseCategories);
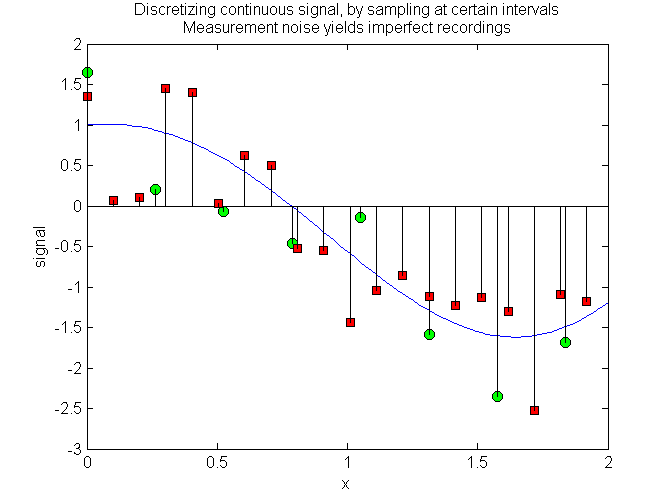
%% stem用于画离散的数据序列
x = linspace(0,2,100);
x1 = x(1:13:end);
x2 = x(1:5:end);
y = exp(.3*x).*cos(-2*x);
yy = round(rand(1,length(x1)));yy(find(yy==0))=-1;
y1 = exp(.3*x1).*cos(-2*x1)+yy.*rand(1,length(x1));
yy = round(rand(1,length(x2)));yy(find(yy==0))=-1;
y2 = exp(.3*x2).*cos(-2*x2)+yy.*rand(1,length(x2));
plot(x,y); hold on;
h1 = stem(x1,y1);
h2 = stem(x2,y2);
% Choose marker size and style of your choice
set(h1,'MarkerFaceColor','green','Marker','o','Markersize',7,'Color',[0 0 0]);
set(h2,'MarkerFaceColor','red','Marker','square','Color',[0 0 0]);
xlabel('x');ylabel('signal');
title({'Discretizing continuous signal, by sampling at certain intervals','Measurement noise yields imperfect recordings'});
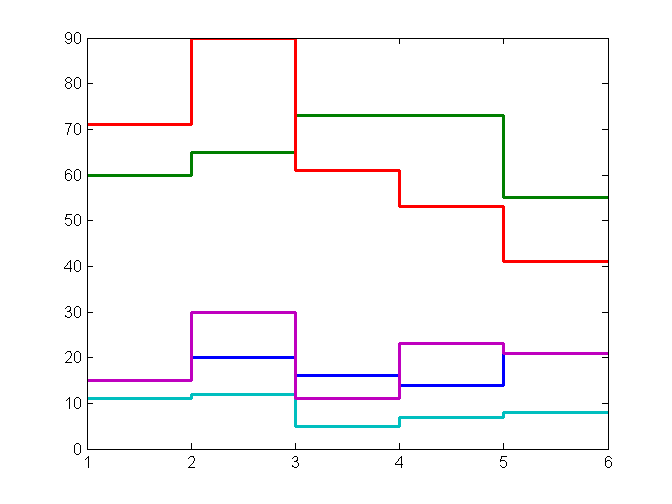
load algoResultsData.mat
h=stairs([MethodPerformanceNumbers nan(5,1)]');
legendMatrix = {'Fresh Tissue','FFPE','Blood','DNA','Simulated'};
for i = 1:5;
set(h(i),'linewidth',2); % thicken the lines
% add total # of samples in this category tolegend
legendMatrix{i} = [legendMatrix{i} ', Total# = ' num2str(CategoryTotals(i))];
end
这篇只是简单的用了三个函数。另外还有pie3,scatter,bar, plot, plotyy这些函数,大家可以去help里面学习下。
另外补充下,在matlab官网看到一个函数pie3s,很漂亮的图,
pie3s([2 4 3 5],'Explode',[0 1 1 0],'Labels',{'North','South','East','West'})
pie3s 代码:
function pie3s( varargin )
%PIE3S: 3-D pie chart with added shininess
%
% PIE3S(X) draws a 3-D pie plot of the data in the vector X. The values
% in X are normalized via X/SUM(X) to determine the area of each slice of
% pie. If SUM(X) <= 1.0, the values in X directly specify the area of
% the pie slices. Only a partial pie will be drawn if SUM(X) < 1.
%
% PIE3S(X,PARAM,VALUE,...) allows additional options to be set. PARAM
% must be one of:
% 'Explode' Specify which elements of the pie should be pulled out.
% The corresponding value must be a vector of ones and
% zeros with size equal to X.
% 'Labels' Specify labels for each pie slice. The corresponding
% value must be a cell array of strings with one string
% per elemet of X.
% 'Bevel' Set the bevelling of the 3D surface. The corresponding
% value must be one of:
% 'In': rounded concave bevel
% 'Out': rounded convex bevel [default]
% 'Flat': straight bevel
% 'Step': straight bevel
% 'Elliptical': a broad ellipse of a bevel
% 'None': no bevel
%
% PIE3S(AX,...) plots into axes AX instead of GCA.
%
% Examples:
% >> pie3s([2 4 3 5],'Explode',[0 1 1 0],'Labels',{'North','South','East','West'})
%
% >> pie3s([2 4 3 5],'Bevel','In','Explode',[0 1 0 0])
%
% >> pie3s([2 4 3 5],'Bevel','Elliptical','Explode',[0 0 0 1])
% >> legend('North','South','East','West')
%
% See also: PIE, PIE3.
% Copyright 2009-2010 The MathWorks Inc.
% $Revision: 40$
% $Date: 2010-05-18$
% Check arguments
error( nargchk( 1, inf, nargin ) );
% Strip off the axes if present
if isa( varargin{1}, 'matlab.graphics.axis.Axes' ) ...
|| (isa( varargin{1}, 'double' ) && isscalar( varargin{1} ) ...
&& ishandle( varargin{1} ) ...
&& strcmp( get( varargin{1}, 'Type' ), 'axes' ))
axh = varargin{1};
varargin(1) = [];
else
axh = gca();
end
% We always clear the axes to avoid problems with lighting and shading
cla( axh );
% Get the X vector
if isempty( varargin ) || ~isnumeric( varargin{1} ) || ~all( varargin{1}>0 )
error( 'PIE3S:BadData', 'Input vector X must be a vector of positive numbers' );
else
data = varargin{1};
varargin(1) = [];
end
% Normalise data
if sum( data ) > 1
data = data / sum( data );
end
% Set some defaults for the optional parameters
bevelType = 'Out';
txtLabels = repmat( {''}, size( data ) );
explode = false( size( data ) );
bite = false( size( data ) );
% Now parse the rest as parameter-value pairs
if ~isempty( varargin );
params = varargin(1:2:end);
values = varargin(2:2:end);
if numel( params ) ~= numel( values )
error( 'PIE3S:BadSyntax', 'Optional inputs must be specified as parameter, value pairs.' );
end
if any( ~cellfun( 'isclass', params, 'char' ) )
error( 'PIE3S:BadParameter', 'Optional parameter names must be character arrays' );
end
for ii=1:numel( params )
switch upper( params{ii} )
case 'LABELS'
txtLabels = values{ii};
case 'BEVEL'
bevelType = values{ii};
case 'EXPLODE'
explode = (values{ii} ~= 0);
case 'BITE'
bite = (values{ii} ~= 0);
otherwise
error( 'PIE3S:BadOption', 'Optional parameter ''%s'' was not recognized', params{ii} );
end
end
end
% Check whether we're over-plotting
bgcol = get( ancestor( axh, 'figure' ), 'Color' );
% OK, let's do some plotting!
theta0 = pi/2;
maxpts = 360;
height = .35;
bevelSize = 0.1;
num_z_levels = 20;
% Set into zbuffer to avoid RGB warnings
set( ancestor( axh, 'figure' ), 'Renderer', 'zbuffer' );
for ii=1:length(data)
n = max(1,ceil(maxpts*data(ii)));
myTheta = 2*pi * linspace(0,data(ii),n+1)';
if bite(ii)
[r,theta] = takeBite( myTheta );
theta = theta + theta0;
else
r = [0;ones(n+1,1);0];
thetaMean = mean( myTheta );
theta = theta0 + [thetaMean;myTheta;thetaMean];
end
[xtext,ytext] = pol2cart(theta0 + data(ii)*pi,1.25);
% Create the outer coords
slice_z = linspace(0,height,num_z_levels);
xx = zeros( numel( theta ), num_z_levels );
yy = zeros( size( xx ) );
zz = zeros( size( xx ) );
for jj=1:num_z_levels
[xx(:,jj),yy(:,jj),zz(:,jj)] = iCreateSlice( height, bevelSize, bevelType, theta, r, slice_z(jj) );
end
if explode(ii)
[xexplode,yexplode] = pol2cart(theta0 + data(ii)*pi,.1);
xtext = xtext + xexplode;
ytext = ytext + yexplode;
xx = xx + xexplode;
yy = yy + yexplode;
end
theta0 = max(theta);
if data(ii)<.01,
lab = '< 1';
else
lab = int2str(round(data(ii)*100));
end
xx0 = xx(1,1)*ones(size(xx,1),1);
yy0 = yy(1,1)*ones(size(yy,1),1);
zz0 = zz(1,1)*ones(size(zz,1),1);
zz1 = zz(1,end)*ones(size(zz,1),1);
colour = ii*ones( size(xx,1), size(xx,2)+2 );
% Create the patches
pieSegment = surface( ...
[xx0,xx,xx0], ...
[yy0,yy,yy0], ...
[zz0,zz,zz1], ...
colour, ...
'Tag', 'Pie3S:Segment', ...
'Parent', axh);
% Creat some shadows
slice = round(num_z_levels/2);
sx = xx(:,slice);
sy = yy(:,slice);
sz = -0.25*ones( size( xx, 1 ), 1 );
shadows = iAddShadow( axh, sx, sy, sz, bgcol );
% position text so that labels near the front don't overlap the patches
z = 0.8 * height * ones(size(xtext));
if ~isempty(txtLabels)
label = text( xtext, ytext, z, txtLabels{ii}, ...
'FontSize', 12, ...
'FontWeight', 'bold', ...
'HorizontalAlignment','center', ...
'Parent', axh, ...
'Clipping', 'off', ...
'Tag', 'Pie3S:Label' );
else
label = text( xtext, ytext, z, [lab,'%'], ...
'FontSize', 12, ...
'FontWeight', 'bold', ...
'HorizontalAlignment','center', ...
'Parent', axh, ...
'Clipping', 'off', ...
'Tag', 'Pie3S:Label' );
end
end
% Set the lighting etc to get a nice visual effect
maxNum = max( 2, numel( data ) );
set( axh, 'CLim', [1 maxNum] );
axis( axh, 'off', 'image', [-1.2 1.2 -1.2 1.2] )
view( axh, [-30 45] )
lighting( 'phong' )
shading( 'flat' )
camlight( 'left' )
% light( 'Parent', axh, 'Position', [40 40 15], 'Style', 'local' );
%-------------------------------------------------------------------------%
function [x,y,z] = iCreateSlice( height, bevel, type, theta, r, z_in )
%iCreateSlice: calculate the X, Y and Z arrays for a single slice through
%one pie segment. This includes bevelling
assert( isscalar( z_in ) );
assert( z_in>=0 && z_in<=height );
z = z_in*ones(size(theta));
if strcmpi( type, 'Elliptical' )
% Special case for elliptical
zRel = (2*z_in - height) / height;
ratio = (1 - zRel^2);
[x,y] = pol2cart(theta,ratio*r);
else
% The others all work the same way, just adding a little decoration
% near to the egdes
if z_in>=bevel && z_in<=(height-bevel)
% On the full size section
[x,y] = pol2cart(theta,r);
else
if z_in>(height-bevel)
z_in = height-z_in;
end
switch upper( type )
case 'IN'
ratio = sqrt(bevel^2 - z_in^2);
case 'OUT'
ratio = bevel - sqrt(bevel^2 - (bevel-z_in)^2);
case 'FLAT'
ratio = bevel - z_in;
case 'STEP'
ratio = bevel/2;
case 'NONE'
ratio = 0;
otherwise
error( id('BadBevelType'), 'BevelType must be one of: ''none'', ''flat'', ''in'', ''out''' );
end
[x,y] = pol2cart(theta,(1-2*ratio)*r);
[x0,y0] = pol2cart(theta(1),ratio);
x = x + x0;
y = y + y0;
end
end
%-------------------------------------------------------------------------%
function patches = iAddShadow( ax, x, y, z, bgcol )
% Create some grey patches below a pie segment
num_patches = 4;
scaling = 1.02;
patches = -1*ones( 1, num_patches );
shade = 0.25;
mid_x = (max( x )+min( x ))/2;
mid_y = (max( y )+min( y ))/2;
% Move the initial shadow in a bit
x = (x-mid_x)/(scaling^2) + mid_x;
y = (y-mid_y)/(scaling^2) + mid_y;
% plot3( mid_x, mid_y, mean(z), 'r+' )
N = numel( x );
for ii=1:num_patches
patches(ii) = patch( ...
'XData', x, ...
'YData', y, ...
'ZData', z, ...
'FaceVertexCData', repmat(0.7,[N,3]), ...
'EdgeColor', shade*bgcol, ...
'AmbientStrength', shade*bgcol(1), ...
'Tag', 'Pie3S:Shadow', ...
'Parent', ax);
% Exclude it from the legend
set(get(get(patches(ii),'Annotation'),'LegendInformation'),'IconDisplayStyle','off');
% Now blur outwards
shade = 1 - (1-shade)*0.6;
x = (x-mid_x)*scaling + mid_x;
y = (y-mid_y)*scaling + mid_y;
z = z - 0.01;
end
%-------------------------------------------------------------------------%
function [r,theta] = takeBite( theta )
%takeBite: take a bite out of a pie slice
%
% [r,theta] = takeBite(theta)
% Make bite
r = ones( size( theta ) );
% Create arc
thetaRange = (max(theta)-min(theta));
theta0 = min(theta) + thetaRange/2;
ratio = zeros(numel(theta),1);
if thetaRange > pi/4
% Too big, so select just the centre pi/4
startIdx = find( theta0 - theta < pi/8, 1, 'first' );
endIdx = find( theta - theta0 < pi/8, 1, 'last' );
thetaRange = theta(endIdx) - theta(startIdx);
else
startIdx = 1;
endIdx = numel(theta);
end
biteDepth = 0.7*thetaRange;
numAngles = endIdx - startIdx + 1;
myRatio = linspace( -0.65, 0.65, numAngles )';
myRatio = (1-myRatio.^2);
rBite = biteDepth * myRatio;
rBite = rBite - min(rBite);
ratio(startIdx:endIdx) = rBite;
% Teeth marks
teethDepth = 0.05;
edges = linspace( -1, 1, 7 )';
spacing = mean( diff( edges ) );
positions = linspace( -1, 1, numAngles )';
dists = inf( numel( positions ), 1 );
for ii=1:numel( edges )
dists = min( dists, abs(positions-edges(ii))/spacing );
end
rTeeth = teethDepth*sqrt(dists);
ratio(startIdx:endIdx) = ratio(startIdx:endIdx) + rTeeth;
% Add zero point
r = [0;r-ratio;0];
theta = [theta0;theta;theta0];