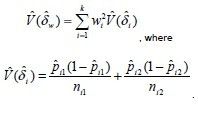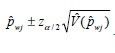Reading "Using SAS/IML to Generate Weighted Chi-Square Statistics"
这篇文章逻辑性很强,脉络清晰,这样的思维能力就是所谓的逻辑了吧.
文章将计算加权卡方统计量分作12个小环节,具体如下:
1.设定矩阵
假设存在一药物作用的患者样本,它有K(1,2,...,K)个观测,两种(i,j)处理,,将所有观测数据写成矩阵形式,就可以得到一个K*2的矩阵
2.计数
要进行卡方统计量的计算,我们首先需要计算行计数和,进而来计算权重,行计数和公式为:
3.比率估计
计算患者药物反应比率的点估计,因为有两种药物处理,所以存在两种处理对应的患者反应情况,再结合K个患者,所以该比率估计也是一个K*2的矩阵
4.加权比率
计算每种处理患者反应的加权比率估计,在(3)计算的比率估计基础上考虑两种处理的权重,重新计算得到的比率就是加权估计比率
iml中表达为:pbar= (n#phat)[,+]/ndot;
5.权重的计算
我们知道标准的Cochran-Mantel-Haenszel的调和平均公式为:
这个公式比较复杂,可以分别计算分子分母的值,然后再进行除法运算,这样在条理可以很清楚看出:
wnum = n[,#]/ndot;
wden = wnum[+,];
w = wnum/wden;
6.每种处理考虑权重的比率估计,
需要区别的是,前面考虑的权重都是单独的观测,这里是对两种处理的进行加权比率估计,得到的结果应是一个1*2的矩阵.
数学表达式为:
iml表达为:pwt = (w#phat)[+,];
7.计算pwt 估计的方差,pwt方差的计算公式为:
iml中的计算代码为:varpwt = (w#w#phat#(1-phat)/n)[+,];
8.计算两种处理间比率差值,显然其差异为:
由于两种处理在矩阵中就是两列,所起该差值就是两列的差值,iml中计算可以简单的表达为:
deltahat = phat[,2] – phat[,1];
上式计算的差值是矩阵中对应原始的差值,还需要计算两组整体的比率差值:
iml中的表达为:deltawt = (w # deltahat) [+,];
9.计算(8)中估计的方差,我们有方差公式:
iml中写成:
vardeltai = (phat#(1-phat) / n ) [,+];
vardeltaw = (w # w # vardeltai ) [+,];
10.计算pwt的置信区间
数学表达;
iml表达:
pwtcil = pwt – 1.96 * sqrt(varpwt);
pwtciu = pwt + 1.96 * sqrt(varpwt);
11.计算deltawt的置信区间
数学表达:
iml表达:
delwcil = deltawt – 1.96* sqrt(vardeltaw);
delwciu = deltawt + 1.96* sqrt(vardeltaw);
12.两种处理的差异性的假设检验,计算卡方值
iml表达:
chictmp = ((n[,#] # deltahat) / ndot ) [+,];
chicnum = chictmp ** 2;
chicden = (n[,#] # pbar # (1-pbar) / ndot ) [+,];
chic2 = chicnum/chicden;
13计算P值
从服从卡方分布的卡方统计量很容易计算得到相应P值:
pchic2 = 1 – probchi(chic2, 1);
14.整理代码如下
data Matrix;
input tx stratum x n;
datalines;
1 1 140 450
1 2 50 170
1 3 60 200
2 1 180 440
2 2 90 200
2 3 60 180
;
run;
%let alpha=0.05;
proc iml;
reset print; /* sent output to window */
use Matrix; /* specify dataset to read in */
read all var {x} into x1 where (tx = 1); /* define matrix x1 from var x*/
read all var {x} into x2 where (tx = 2); /* and same for matrix x2 */
read all var {n} into n1 where (tx = 1); /* likewise for matrix n1 */
read all var {n} into n2 where (tx = 2); /* finally, matrix n2 */
x = x1 || x2; /* Catenate x1 and x2 to get X */
n = n1 || n2; /* Catenate n1 and n2 to get N */
ndot = n[,+]; /* All statements below were */
phat = x/n; /* previously explained above */
pbar = (n#phat)[,+] / ndot;
wnum = n[,#] / ndot;
wden = wnum[+,];
w = wnum / wden;
pwt = (w # phat)[+,];
varpwt = (w # w # phat # (1-phat) / n)[+,];
deltahat = phat[,2] – phat[,1];
deltawt = (w # deltahat) [+,];
vardeltai = (phat # (1-phat) / n ) [,+];
vardeltaw = (w # w # vardeltai ) [+,];
%let uppera = %sysevalf(1.0-&alpha/2);
zdot5 = probit(&uppera);
pwtcil = pwt - zdot5 * sqrt(varpwt);
pwtciu = pwt + zdot5 * sqrt(varpwt);
delwcil = deltawt - zdot5 * sqrt(vardeltaw);
delwciu = deltawt + zdot5 * sqrt(vardeltaw);
chictmp = ( (n[,#] # deltahat) / ndot ) [+,];
chicnum = chictmp ** 2;
chicden = (n[,#] # pbar # (1-pbar) / ndot ) [+,];
chic2 = chicnum/chicden;
pchic2 = 1 – probchi(chic2, 1);
quit;
详见:<Using SAS/IML to Generate Weighted Chi-Square Statistics>