一、需求
所要爬取的数据库是CKB数据库
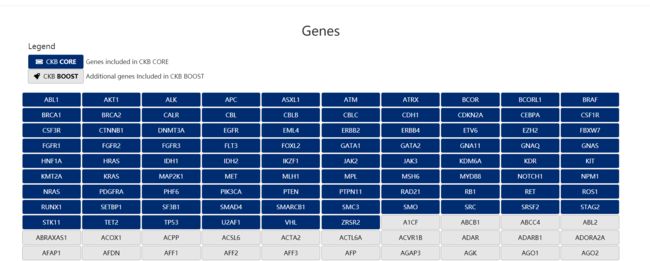
点击进入Genes页面
蓝色的gene名的部分是数据库公布的,可获得的信息
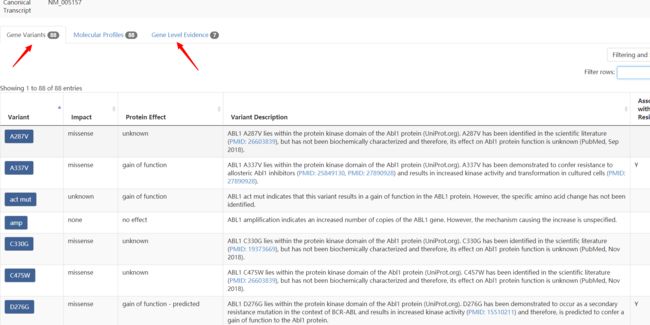
随便点击一个,比如ABL1
可以看到对应的页面下,存在箭头所指的两个表格
而,需求就是将所有蓝色部分的gene所对应的这两个表格爬取下来
二、所使用的技术路线
1.路线
这里所使用的方法是requests-BeautifulSoup4
requests.get()获取页面内容
BeautifulSoup4解析页面
2.分析
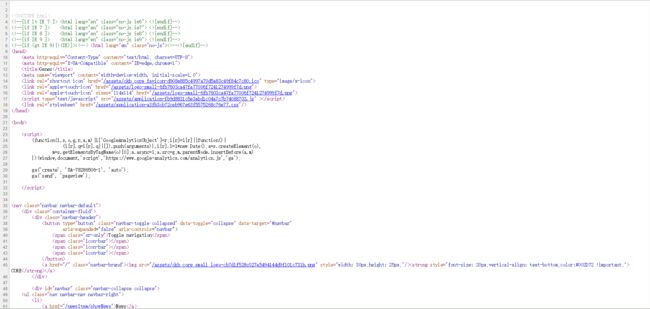
首先看一下下图的网页源代码
先简单的找到比如说ABL1相关的信息
可以看出来ABL1是由
和两个标签包围的
其他的基因也是这样
比如
可以看出来class信息是一样的,而href信息可以获得每个gene对用的页面链接信息
因此,可以用soup.find_all,将name设为"a",attrs设为"btn btn-default btn-gene btn-block"来获得所有gene的信息
将gene 和对应的链接以字典形式存储
然后,可以看一下某个gene页面,比如ABL1
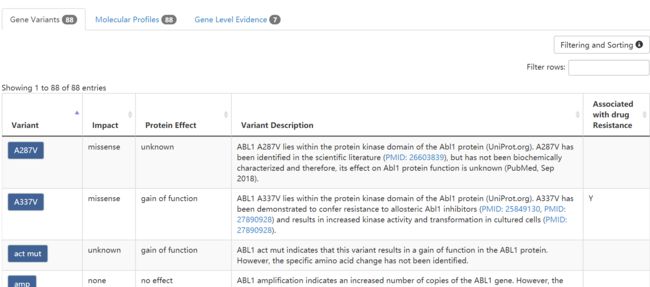
先看第一个表,Gene Variants
第二个表,Gene Level Evidence
看一下源代码
先找到和第一个表格相关的地方
基本上,可以看出来每个Variant对应的信息被包括在tbody以及两个子标签tr和td中
同理找一下第二个表格相关的信息即可
三、全代码
import requests
import bs4
from bs4 import BeautifulSoup
def getHTMLText(url):
try:
r = requests.get(url,timeout = 30)
r.raise_for_status()
r.encoding = r.apparent_encoding
return r.text
except:
return ""
def get_gene_id(url):
html = getHTMLText(url)
soup = BeautifulSoup(html,'html.parser')
gene_id_dict = {}
for a in soup.find_all(name = "a",attrs = "btn btn-default btn-gene btn-block"):
gene_name = a.string.replace("\n","").replace(" ","")
ID = a.attrs['href']
gene_id = "https://ckb.jax.org" + ID
gene_id_dict[gene_name] = gene_id
return gene_id_dict
def gene_variant_list(alist,html):
soup = BeautifulSoup(html,'html.parser')
for tr in soup.find_all("tbody")[1].children:
if isinstance(tr,bs4.element.Tag):
var_des = ''
tds = tr("td")
variant = tds[0].a.string.replace(" ",'').replace("\n",'')
for string in tds[3].strings:
var_des += string
var_des = var_des.replace("\n","")
alist.append([variant,tds[1].string,tds[2].string,var_des,tds[4].string])
def gene_level_evidence_list(blist,html):
soup = BeautifulSoup(html,'html.parser')
num = 0
for name in soup.find_all(name = "a",attrs = "btn btn-default btn-wrap btn-therapy"):
num += 1
count = num*8
eight_list = []
for k in range(count):
string_extract = ""
if (k+1)%8 == 1:
if k != 0 :
blist.append([eight_list[0],eight_list[1],eight_list[2],eight_list[3],eight_list[4],eight_list[5],eight_list[6],eight_list[7]])
eight_list = []
for i in soup.find_all("td")[-count:][k]:
a = i.string.replace("\n","") #string extract
aL = a.split(" ")
aL = list(filter(None,aL))
a = " ".join(aL)
string_extract += " "+ a #delete blank and get string
eight_list.append(string_extract)
if eight_list:
blist.append([eight_list[0],eight_list[1],eight_list[2],eight_list[3],eight_list[4],eight_list[5],eight_list[6],eight_list[7]])
def print_gene_variant_list(alist,gene):
num = len(alist)
for i in range (num):
u = alist[i]
print (gene,u[0],u[1],u[2],u[3],u[4],sep="\t")
def print_gene_level_evidence_list(blist,gene):
num = len(blist)
if num :
for i in range (num):
m = blist[i]
print (gene,m[0],m[1],m[2],m[3],m[4],m[5],m[6],m[7],sep = "\t")
else:
if num == 0:
print(gene)
def main():
url = "https://ckb.jax.org/gene/grid"
gDict = get_gene_id(url)
print("{:^10}\t{:^6}\t{:^10}\t{:^10}\t{:^10}\t{:^10}".format("Gene_Name","Variant","Impact","Protein_Effect","Variant_Description","Associated_with_drug_resistance"))
for name,ID in gDict.items():
ainfo = []
url = ID
gene = name
html = getHTMLText(url)
gene_variant_list(ainfo,html)
print_gene_variant_list(ainfo,gene)
print("{:^10}\t{:^10}\t{:^10}\t{:^10}\t{:^10}\t{:^10}\t{:^10}\t{:^10}\t{:^10}".format("Gene_Name","Molecular_Profile","Indication/Tumour_type","Response_Type","Therapy_Name","Approval_Status","Evidence_Type","Efficacy_Evidence","References"))
for name,ID in gDict.items():
binfo = []
url = ID
gene = name
html = getHTMLText(url)
gene_level_evidence_list(binfo,html)
print_gene_level_evidence_list(binfo,gene)
main()
cat craw_3.xls |tr -d "\r" >new.xls