阅读文献
Couvin D, Bernheim A, Toffanonioche C, et al. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins.[J]. Nucleic Acids Research, 2018, 46(Web Server issue):W246-W251.
CRISPR-Cas系统
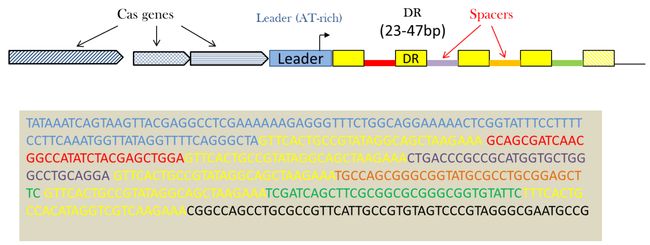
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPRs) are specific structures found in many prokaryotic genomes that show characteristics of both tandem and interspaced repeats. They have been described in a wide range of prokaryotes, including the majority of Archae and many Eubacteria. A CRISPR locus is characterized by:
- Repeats and Spacers : A CRISPR is a succession of 23-47bp sequences called repeats separated by unique sequences of a similar length (spacers). Sometimes, at one end of the CRISPR, the repeat is not totally conserved, it is called degenerate repeat.
- A leader sequence : the CRISPR locus is generally flanked on one side by an AT-rich leader sequence of 100-350 bp, acting as a promoter for the pre-crRNA synthesis.
Together with a set of genes called cas for “CRISPR-associated”, they constitute an immune system.
- Cluster of cas genes : CRISPR-associated genes are genes found closely linked to the repetitive sequences.
Repeats and Spacers
In a given strain several CRISPRs can be found with a single or different repeat sequences but only one of each kind is associated with the cas genes. The spacers in the different CRISPRs are different.
The unique sequences or spacers correspond mostly to fragments of foreign DNA, ie. viruses, plasmids or mobile genetic elements.
Cas Genes
Several genes called cas for CRISPR-associated are found in the vicinity of CRISPRs and perform the three different functions of the immune system: adaptation, crRNA maturation and interference. Their number varies from one type to another. Phylogenetic studies performed on the CAS protein suggest that CRISPRs are acquired by horizontal transfer. This is further shown by their presence on megaplasmids.
Leader sequence
CRISPR loci are transcribed into a pre-crRNA from the leader acting as a promoter. This precursor is then matured into small crRNA that play a role in the targeting and destruction of homologous foreign sequences.
CRISPR-Cas系统: 是在细菌(>50%)和古菌(>90%)中广泛存在的对外源病毒和质粒具有特异性抗性的、 获得性免疫系统。CRISPR 由短的高度保守的重复序列(repeat)和各不相同的间隔序列(spacer)组成 。Repeat多具有回文结构。Spacer与外源DNA( 如质粒或病毒) 同源。由CRISPR转录加工形成的crRNA,可通过与Cas功能蛋白形成复合物,特异识别和消除入侵细胞的外源质粒或病毒。
CRISPR are repeat arrays found in the DNA of many bacteria and archaea. The name is an acronym for Clustered Regularly Interspaced Short Palindromic Repeats.
The repeats or DR, ranging in size from 23 to 47 base pairs, are separated by spacers of similar length. Repeats often show some dyad symmetry but are not truly palindromic. Spacers are usually unique in a genome. They match sequences in genomes of phage, plasmid or mobile genetic elements. Inside a species, the CRISPR repeat array may show polymorphism.
Cas genes stand for CRISPR-associated genes. Together with the CRISPR array they constitute the CRISPR-Cas defense mechanism. Cas function as clusters of 3 to more than 10 genes and can be distributed into 6 types (I to VI) and more than 30 subtypes.
CRISPRCasFinder可以鉴定CRISPR序列和Cas蛋白。 该软件包括:(i)改进的CRISPR序列检测工具,促进基于评级系统的专业验证机制;(ii)预测CRISPR方向;(iii)更新Cas蛋白检测和分型工具以匹配这些系统的最新分类方案。 CRISPRCasFinder既可以在线使用,也可以作为与Linux操作系统兼容的独立工具使用。 该程序使用的所有第三方软件包都是免费提供的。
软件安装
官网链接:https://crisprcas.i2bc.paris-saclay.fr.
CRISPRCasFinder程序可以在用户提交的序列数据中轻松检测CRISPR序列和cas基因(允许序列高达50 Mo,否则下载独立程序)。 该软件是CRISPRFinder软件的更新版,具有改进的特异性和CRISPR方向的指示。 MacSyFinder用于鉴定cas基因、CRISPR-Cas类型和亚型。
这个软件的依赖特别多,当我看到下面的说明书时我就在使用conda或者docker容器去安装,而不是自己没个去手动安装。
如果手动安装依赖需要怎样?
大概需要像下面一样,看看都头晕~
mkdir -p ${SINGULARITY_ROOTFS}/usr/local/src/CRISPRCasFinder
cp CRISPRCasFinder.singularity.patch ${SINGULARITY_ROOTFS}/usr/local/src/CRISPRCasFinder/
export DEBIAN_FRONTEND=noninteractiveapt-get updateapt-get install -y apt-utils zlib1g-dev make gcc# dash is too restrictedln -nsf /bin/bash /bin/sh
# to be runnable on tars @ Institut Pasteur
mkdir /pasteur
apt-get update -y
apt-get install -y curl default-jre python perl parallel cpanminus patch wget unzip
###################
# Bioinfo package #
###################
apt-get install -y \
hmmer \
emboss emboss-lib \
ncbi-blast+ \
bioperl \
bioperl-run \
libdatetime-perl \
libxml-simple-perl \
libdigest-md5-perl \
clustalw \
muscle \
prodigal \
aragorn \
infernal \
cd /usr/bin
ln -s clustalw2 clustalw2
cd /
cpanm Try::Tiny
cpanm Test::Most
cpanm JSON::Parse
cpanm Date::Calc
cpanm Class::Struct
cpanm Bio::DB::Fasta
cpanm File::Copy
cpanm Bio::Seq Bio::SeqIO
cpanm --force Bio::Tools::Run::Alignment::Clustalw
cpanm --force Bio::Tools::Run::Alignment::Muscle
prefix="/usr/local"
##########
# vmatch #
##########
PN="vmatch"
PV="2.3.0"
P="${PN}-${PV}"
P_SRC=${prefix}/src/${PN}
mkdir -p ${prefix}/src/vmatch
cd ${prefix}/src/vmatch
distribution='Linux_x86_64'
vmatch="${PN}-${PV}-${distribution}-64bit"
vmatch_url="http://vmatch.de/distributions/${vmatch}.tar.gz"
curl -L -O --silent "${vmatch_url}"
tar -zxf ${vmatch}.tar.gz
cd ${vmatch}
gcc -Wall -Werror -fPIC -O3 -shared SELECT/sel392.c -m64 -o sel392v2.so
# copy the shared library in LD_LIBRARY_PATH
install -m 0775 sel392v2.so /.singularity.d/libs/sel392v2.so
cd /.singularity.d/libs/
ln -s sel392v2.so sel392.so
cd ${prefix}/src/${PN}/${vmatch}
install -m 0775 vmatch ${prefix}/bin/vmatch2
install -m 0775 vsubseqselect ${prefix}/bin/vsubseqselect2
install -m 0775 mkvtree ${prefix}/bin/mkvtree2
cd /
###############
# macsyfinder #
###############
PN="macsyfinder"
PV="1.0.5"
P="${PN}-${PV}"
P_SRC=${prefix}/src/${PN}
mkdir -p ${prefix}/src/${PN}
cd ${prefix}/src/${PN}
macsyfinder_url="https://dl.bintray.com/gem-pasteur/MacSyFinder/${P}.tar.gz"
curl -L -O --silent "${macsyfinder_url}"
tar -xzf ${P}.tar.gz
cd ${P}
python setup.py build
python setup.py install
cd /
#######################
# prokka dependencies #
#######################
###########
# signalp #
###########
# Cannot be installed due to Licensing problem.
###########
# tbl2asn #
###########
# trusty package ncbi-tools-bin provide a too old tbl2asn
PN="tbl2asn"
PV="1.12"
P="${PN}-${PV}"
P_SRC=${prefix}/src/${PN}
mkdir -p ${P_SRC}
cd ${prefix}/src/tbl2asn
tbl2asn_url="ftp://ftp.ncbi.nih.gov/toolbox/ncbi_tools/converters/by_program/${PN}/linux64.${PN}.gz"
wget "${tbl2asn_url}"
gunzip linux64.tbl2asn.gz
install -m 0755 linux64.tbl2asn ${prefix}/bin/${PN}
##########
# prokka #
##########
PN="prokka"
PV="1.12"
P="${PN}-${PV}"
P_SRC=${prefix}/src/${PN}
mkdir -p ${P_SRC}
cd ${P_SRC}
prokka_url="http://www.vicbioinformatics.com/${P}.tar.gz"
curl -L -O --silent "${prokka_url}"
tar -xzf ${P}.tar.gz
cd ${P}
prokka_data=${prefix}/share/${PN}
prokka_db=${prokka_data}/db
test -d ${prokka_db} || mkdir -p ${prokka_db}
# copy database
cp -pr db/* ${prokka_db}
# tell prokka where to find its tools and db once installed
sed -i -e "s|my \$BINDIR.*|my \$BINDIR=\"${prefix}/libexec/prokka\";|" \
-e "s|my \$DBDIR.*|my \$DBDIR=\"${prokka_db}\";|" \
bin/prokka
for bin in bin/*;
do
install -m 0755 ${bin} ${prefix}/bin/
done
# install prokka binaries
test -d ${prefix}/libexec/${PN} || mkdir -p ${prefix}/libexec/${PN}
for p in binaries/linux/*;
do
install -m 0755 ${p} ${prefix}/libexec/${PN}
done
# parallel is installed via packet manager
install -m 0755 binaries/common/minced ${prefix}/libexec/${PN}/
install -m 0644 binaries/common/minced.jar ${prefix}/libexec/${PN}/
# setup prokka db
prokka_cmd="${prefix}/bin/${PN}"
${prokka_cmd} --setupdb
cd /
###################
# CRISPRCasFinder #
###################
PN="CRISPRCasFinder"
PV="4.2.18"
P="${PN}-${PV}"
test -d "${prefix}/src/${PN}" || mkdir -p "${prefix}/src/${PN}"
cd "${prefix}/src/${PN}"
cripsr_cas_url="https://github.com/bneron/${PN}/archive/release-${PV}.tar.gz"
curl -L -o "${PN}.tar.gz" --silent "${cripsr_cas_url}"
tar -xzf "${PN}.tar.gz" --strip-component 1
crispr_data="${prefix}/share/${PN}"
test -d "${crispr_data}" || mkdir "${crispr_data}"
patch CRISPRCasFinder.pl CRISPRCasFinder.patch
patch CRISPRCasFinder.pl singularity/CRISPRCasFinder.singularity.patch
install -m 0755 CRISPRCasFinder.pl ${prefix}/bin/CRISPRCasFinder
install -m 0644 supplementary_files/crispr.css ${crispr_data}
install -m 0644 supplementary_files/Repeat_List.csv ${crispr_data}
install -m 0644 supplementary_files/CRISPR_crisprdb.csv ${crispr_data}
install -m 0644 supplementary_files/repeatDirection.tsv ${crispr_data}
#############
# CasFinder #
#############
# use the CasFinder distributed with CRISPRCasFinder
cas_data="${prefix}/share/macsyfinder/"
# remove profiles and definitions packaged with macsyfinder
rm -Rf "${cas_data}DEF"
rm -Rf "${cas_data}profiles"
# install cas profiles and definition packaged with CRISPRCasFinder
cp -r CasFinder-2.0.2 ${cas_data}
cd /
Vmatch version 2.3.0 (http://www.vmatch.de/download.html)
EMBOSS version 5.0.0 or upper (http://emboss.sourceforge.net/)
Prodigal version 2.6.3 (https://github.com/hyattpd/Prodigal)
MacSyFinder version 1.0.5 (https://github.com/gem-pasteur/macsyfnder)
Muscle (version 3.8.31) (http://www.drive5.com/muscle)
Perl (https://www.perl.org/). The installer_MAC.sh will install perl5.
BioPerl version 1.6.2 or upper (http://bioperl.org/)
installer_MAC.sh will also install prokka-1.12 and tbl2asn
一共九个依赖软件,一个安装出现问题都会导致软件无法正常安装。
创建一个自己的docker镜像
很悲惨,我没有在doker中找到相应的镜像,所以我尝试在docker中自己建一个,但是俺在bioperl模块那里卡住了,总是安装不成功,怪我学艺不精了。
使用singularity容器中的镜像
apt-get update && apt-get install -y \
build-essential \
libssl-dev \
uuid-dev \
libgpgme11-dev
apt install wget
export VERSION=1.11 OS=linux ARCH=amd64
cd /tmp
wget https://dl.google.com/go/go1.11.1.linux-amd64.tar.gz
tar -C /usr/local -xzf go1.11.1.linux-amd64.tar.gz
echo 'export GOPATH=${HOME}/go' >> ~/.bashrc
echo 'export PATH=/usr/local/go/bin:${PATH}:${GOPATH}/bin' >> ~/.bashrc
source ~/.bashrc
mkdir -p $GOPATH/src/github.com/sylabs
cd $GOPATH/src/github.com/sylabs
apt install git
git clone https://github.com/sylabs/singularity.git
cd singularity
go get -u -v github.com/golang/dep/cmd/dep
cd $GOPATH/src/github.com/sylabs/singularity
./mconfig
make -C builddir
make -C builddir install
singularity help
### 下面的镜像来自https://www.singularity-hub.org/collections/1625:
singularity pull --name CRISPRCasFinder shub://bneron/CRISPRCasFinder:latest
singularity pull --name CRISPRCasFinder shub://bneron/CRISPRCasFinder:4.2.18
./CRISPRCasFinder -def General -cas -i my_sequence.fasta -keep
参数设置
CRISPR高级参数设置
默认参数的设置能检测到高度同源的重复序列。但还是有需求修改优化某些参数来定义最大重复序列和CRISPR的属性。
- 最小重复序列长度:Minimal Repeat length (默认为23 ; 可调范围1~70)
- 最大重复序列长度:Maximal Repeat length (默认为55 ; 可调范围2~80)
- 重复序列中允许错配数:Allow mismatch between repeats (默认为1; 可设值1/0)
- 最小间隔长度与功能重复序列长度的比值:Minimal Spacers size in function of Repeat size (默认为0.6 ; 可调范围0.1~60)
- 最大间隔长度与功能重复序列长度的比值:Maximal Spacers size in function of Repeat size (默认为2.5 ; 可调范围1.5~60)
- 间隔序列直接的最大相似度:Maximal allowed percentage of similarity between Spacers (默认为60 ; 可调范围1~100)
- 重复序列之间的错配率:Percentage mismatches allowed between Repeats (默认为20 ; 可调范围1~100)
- 平头重复序列的错配率:Percentage mismatches allowed for truncated Repeat (默认为33.3 ; 可调范围1 ~100)
CRISPR其他参数设置
- CRISPR的侧翼序列能够被修饰数量:The size of Flanking regions in base pairs (bp) for each analyzed CRISPR array can be modified (默认100 ; 可调范围10~1000).
- 检测truncated重复序列的方法选择. Mismatches are search in the first half of the repeat flanking the array.
CAS参数设置
- Perform CAS detection有三种严格的等级去识别cas基因。
- General: allows a permissive search (i.e. CAS will be detected whatever the system type or subtype).
- Typing and SubTyping: produce more stringent analyses.
具体细节可以看
MacSyFinder软件文档(http://macsyfinder.readthedocs.io/en/latest/) 。
- The "Unordered" button allows users to perform a search for non-clustered cas genes in unordered or smaller sequences (such as contigs). 该功能需要在
MacSyFinder设置Prodigal软件的参数为"-p meta --db-type unordered" .
可视化结果
The summary displays information on CRISPR arrays and cas gene clusters in the order in which they lie along the chromosome. Direction is the proposed orientation of the CRISPR array (ND is for Not determined) according to the CRISPRDirection program. In Details is shown, in addition, the potential orientation of the CRISPR array based on the AT percentage in 100bp flanking sequences.
"Conservation DR" corresponds to the EBcons (Entropy-Based conservation) of repeats as described in the related manuscript (Couvin et al., NAR 2018).
"Conservation Spacer" indicates the conservation of spacers based on BioPerl's overall percentage identity.
参考文献
- CRISPRFinder : a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007, 35: W52-7
- CRISPRCasFinder : an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018
- MacSyFinder : a program to mine genomes for molecular systems with an application to CRISPR-Cas systems. PloS One 2014 7;9(10):e110726
如果实在有需要请给我发邮件:[email protected];
也可以关注我的公众号:沈梦圆(PandaBiotrainee)