一次完全失败的尝试
COMPLETE FAILURE
以 HTML table 形式呈现芯片数据的好处是可以加上各个公共注释数据库的链接,便于下游分析,而且小巧便捷,用浏览器可以直接打开。
制作这样的 HTML table 的先决条件是要有所用芯片的注释信息,当然制造商一般会提供可导入 R 的文件格式,例如用 read.csv() 就可以加载 Affymetrix 的 csv 文件。
1. 加载包&数据准备
以 Affymetrix HG-U95Av2 芯片为例,随机取15个基因:
library(annotate)
data("sample.ExpressionSet")
igenes <- featureNames(sample.ExpressionSet)[246:260]
2. 准备注释数据
library(hgu95av2.db)
## GenBank ID
gb <- as.character(unlist(lookUp(igenes, "hgu95av2", "ACCNUM")))
gb
# [1] "M57423" "Z70218" "L17328" "S81916" "U63332" "M77235"
# [7] "X98175" "AB019392" "J03071" "D25272" "D63789" "D63789"
# [13] "U19142" "U19147" "X16863"
## LocusLink ID
ll <- as.character(unlist(lookUp(igenes, "hgu95av2", "ENTREZID")))
ll
# [1] "221823" "4330" "9637" "5230" "4287" "6331" "841" "27335"
# [9] NA NA "6846" NA NA NA "2215"
## UniGene ID
ug <- as.character(unlist(lookUp(igenes, "hgu95av2", "UNIGENE")))
ug
# [1] "Hs.169284" "Hs.268515" "Hs.258563" "Hs.78771" "Hs.532632" "Hs.517898"
# [7] "Hs.599762" "Hs.314359" NA NA "Hs.458346" NA
# [13] NA NA "Hs.372679" "Hs.694258" "Hs.736230"
## SwissProt ID 一个探针对应多个UNIPROT ID,保留为list格式
sp <- lookUp(igenes, "hgu95av2", "UNIPROT")
names(sp) <- NULL
sp
# [[1]]
# [1] "P21108"
#
# [[2]]
# [1] "A0A024R1C3" "Q10571"
#
# [[3]]
# [1] "Q9UHY8"
#
# [[4]]
# [1] "P00558" "V9HWF4"
#
# [[5]]
# [1] "P54252" "A0A0A0MS38" "C9JQV6"
#
# [[6]]
# [1] "Q14524" "Q86V90" "H9KVD2" "E9PG18" "E9PHB6"
# [6] "K4DIA1" "A0A0A0MT39"
#
# [[7]]
# [1] "A0A024R3Z8" "Q14790"
#
# [[8]]
# [1] "K7ERF1" "U3LUI4" "K7EK53" "Q9UBQ5"
#
# [[9]]
# [1] NA
#
# [[10]]
# [1] NA
#
# [[11]]
# [1] "Q9UBD3"
#
# [[12]]
# [1] NA
#
# [[13]]
# [1] NA
#
# [[14]]
# [1] NA
#
# [[15]]
# [1] "O75015" "M9MML6" "A0A087WZR4" "A0A087WU90"
需要把 NA 转换为 " ; ",原因嘛,我也说不清....
Because the code for SwissProt IDs will not automatically handle missing data, we have to convert the missing data to an HTML empty cell identifier (" ")
## 写个循环改一下
for(i in 1:length(names)) {
if(i == 9 || i == 10 || i ==12 ||
i ==13 || i == 14)
names[i] <- " "
}
sp
# [[1]]
# [1] "P21108"
#
# [[2]]
# [1] "A0A024R1C3" "Q10571"
#
# [[3]]
# [1] "Q9UHY8"
#
# [[4]]
# [1] "P00558" "V9HWF4"
#
# [[5]]
# [1] "P54252" "A0A0A0MS38" "C9JQV6"
#
# [[6]]
# [1] "Q14524" "Q86V90" "H9KVD2" "E9PG18" "E9PHB6"
# [6] "K4DIA1" "A0A0A0MT39"
#
# [[7]]
# [1] "A0A024R3Z8" "Q14790"
#
# [[8]]
# [1] "K7ERF1" "U3LUI4" "K7EK53" "Q9UBQ5"
#
# [[9]]
# [1] " "
#
# [[10]]
# [1] " "
#
# [[11]]
# [1] "Q9UBD3"
#
# [[12]]
# [1] " "
#
# [[13]]
# [1] " "
#
# [[14]]
# [1] " "
#
# [[15]]
# [1] "O75015" "M9MML6" "A0A087WZR4" "A0A087WU90"
## Gene Name
names <- as.character(unlist(lookUp(igenes, "hgu95av2", "GENENAME")))
names
# [1] "phosphoribosyl pyrophosphate synthetase 1 like 1"
# [2] "MN1 proto-oncogene, transcriptional regulator"
# [3] "fasciculation and elongation protein zeta 2"
# [4] "phosphoglycerate kinase 1"
# [5] "ataxin 3"
# [6] "sodium voltage-gated channel alpha subunit 5"
# [7] "caspase 8"
# [8] "eukaryotic translation initiation factor 3 subunit K"
# [9] NA
# [10] NA
# [11] "X-C motif chemokine ligand 2"
# [12] NA
# [13] NA
# [14] NA
# [15] "Fc fragment of IgG receptor IIIb"
for(i in 1:length(names)) {
if(i == 9 || i == 10 || i ==12 ||
i ==13 || i == 14)
names[i] <- " "
}
names
# [1] "phosphoribosyl pyrophosphate synthetase 1 like 1"
# [2] "MN1 proto-oncogene, transcriptional regulator"
# [3] "fasciculation and elongation protein zeta 2"
# [4] "phosphoglycerate kinase 1"
# [5] "ataxin 3"
# [6] "sodium voltage-gated channel alpha subunit 5"
# [7] "caspase 8"
# [8] "eukaryotic translation initiation factor 3 subunit K"
# [9] " "
# [10] " "
# [11] "X-C motif chemokine ligand 2"
# [12] " "
# [13] " "
# [14] " "
# [15] "Fc fragment of IgG receptor IIIb"
3. 一些相关的值
取前10个基因作为例子:
dat <- exprs(sample.ExpressionSet)[igenes,1:10]
FC <- rowMeans(dat[igenes,1:5]) - rowMeans(dat[igenes,6:10])
pval <- esApply(sample.ExpressionSet[igenes,1:10], 1, function(x) t.test(x[1:5], x[6:10])$p.value)
tstat <- esApply(sample.ExpressionSet[igenes,1:10], 1, function(x) t.test(x[1:5], x[6:10])$statistic)
4. 构建 HTML table
genelist <- list(igenes, ug, ll, gb, sp)
filename <- "Interesting_genes.html"
title <- "An Artificial Set of Interesting Genes"
othernames <- list(names, round(tstat, 2), round(pval, 3), round(FC, 1), round(dat, 2))
head <- c("Probe ID", "UniGene", "LocusLink", "GenBank", "SwissProt", "Gene Name", "t-statistic", "p-value",
"Fold Change", "Sample 1", "Sample 2", "Sample 3", "Sample 4", "Sample 5", "Sample 6",
"Sample 7", "Sample 8", "Sample 9", "Sample 10")
repository <- list("affy", "ug", "en", "gb", "sp")
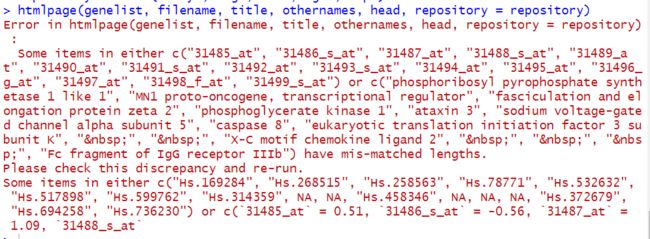
htmlpage(genelist, filename, title, othernames, head, repository = repository)
然而很绝望....
所以是为什么呢,一步一步和作者的代码对照,只能发现
区别主要就是这么个区别,于是只好再操作一波。
for(i in 1:15) {
if(i == 9 || i == 10 || i ==12 ||
i ==13 || i == 14)
ll[i] <- "---"
}
for(i in 1:15) {
if(i == 9 || i == 10 || i ==12 ||
i ==13 || i == 14)
ug[i] <- "---"
}
ll
# [1] "221823" "4330" "9637" "5230" "4287" "6331" "841" "27335" "---"
# [10] "---" "6846" "---" "---" "---" "2215"
ug
# [1] "Hs.169284" "Hs.268515" "Hs.258563" "Hs.78771" "Hs.532632" "Hs.517898"
# [7] "Hs.599762" "Hs.314359" "---" "---" "Hs.458346" "---"
# [13] "---" "---" "Hs.372679" "Hs.694258" "Hs.736230"
还是不行....一毛一样的报错
绝望中打开了作者的 R Script
所以其实都是这样来的吗
那么,如果,按照这些代码从头来一遍会怎样呢?
fine 果然成功了。
而且也没多么好看嘛,作者原话是 nice looking&pretty
“已经折腾了半天就不能白折腾”
现在要开始做一些也许毫无意义的排错。
> gb_t <- as.character(unlist(lookUp(igenes, "hgu95av2", "ACCNUM")))
> gb
[1] "M57423" "Z70218" "L17328" "S81916" "U63332"
[6] "M77235" "X98175" "AB019392" "J03071" "D25272"
[11] "D63789" "D63789" "U19142" "U19147" "X16863"
> gb_t
[1] "M57423" "Z70218" "L17328" "S81916" "U63332"
[6] "M77235" "X98175" "AB019392" "J03071" "D25272"
[11] "D63789" "D63789" "U19142" "U19147" "X16863"
> class(gb)
[1] "character"
> class(gb_t)
[1] "character"
GenBank ID clear
> ll_t <- as.character(unlist(lookUp(igenes, "hgu95av2", "ENTREZID")))
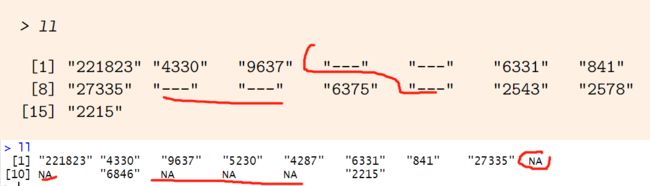
> ll
[1] "221823" "4330" "9637" "---" "---" "6331" "841"
[8] "27335" "---" "---" "6375" "---" "2543" "2578"
[15] "2215"
> ll_t
[1] "221823" "4330" "9637" "5230" "4287" "6331" "841"
[8] "27335" NA NA "6846" NA NA NA
[15] "2215"
> for(i in 1:15) {
+ if(i == 9 || i == 10 || i ==12 ||
+ i ==13 || i == 14)
+ ll_t[i] <- "---"
+ }
> ll_t
[1] "221823" "4330" "9637" "5230" "4287" "6331" "841"
[8] "27335" "---" "---" "6846" "---" "---" "---"
[15] "2215"
> class(ll)
[1] "character"
> class(ll_t)
[1] "character"
LocusLink ID clear
> ug_t <- as.character(unlist(lookUp(igenes, "hgu95av2", "UNIGENE")))
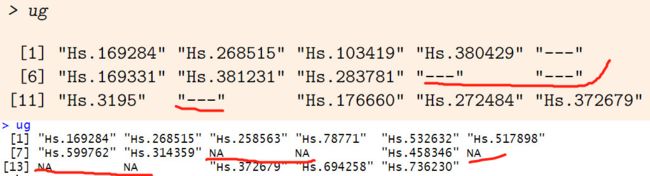
> ug
[1] "Hs.169284" "Hs.268515" "Hs.103419" "Hs.380429" "---"
[6] "Hs.169331" "Hs.381231" "Hs.283781" "---" "---"
[11] "Hs.3195" "---" "Hs.176660" "Hs.272484" "Hs.372679"
> ug_t
[1] "Hs.169284" "Hs.268515" "Hs.258563" "Hs.78771" "Hs.532632"
[6] "Hs.517898" "Hs.599762" "Hs.314359" NA NA
[11] "Hs.458346" NA NA NA "Hs.372679"
[16] "Hs.694258" "Hs.736230"
> for(i in 1:15) {
+ if(i == 9 || i == 10 || i ==12 ||
+ i ==13 || i == 14)
+ ug_t[i] <- "---"
+ }
> ug_t
[1] "Hs.169284" "Hs.268515" "Hs.258563" "Hs.78771" "Hs.532632"
[6] "Hs.517898" "Hs.599762" "Hs.314359" "---" "---"
[11] "Hs.458346" "---" "---" "---" "Hs.372679"
[16] "Hs.694258" "Hs.736230"
> class(ug)
[1] "character"
> class(ug_t)
[1] "character"
UniGene ID clear
> sp_t <- lookUp(igenes, "hgu95av2", "UNIPROT")
> sp_t
$`31485_at`
[1] "P21108"
$`31486_s_at`
[1] "A0A024R1C3" "Q10571"
$`31487_at`
[1] "Q9UHY8"
$`31488_s_at`
[1] "P00558" "V9HWF4"
$`31489_at`
[1] "P54252" "A0A0A0MS38" "C9JQV6"
$`31490_at`
[1] "Q14524" "Q86V90" "H9KVD2" "E9PG18" "E9PHB6"
[6] "K4DIA1" "A0A0A0MT39"
$`31491_s_at`
[1] "A0A024R3Z8" "Q14790"
$`31492_at`
[1] "K7ERF1" "U3LUI4" "K7EK53" "Q9UBQ5"
$`31493_s_at`
[1] NA
$`31494_at`
[1] NA
$`31495_at`
[1] "Q9UBD3"
$`31496_g_at`
[1] NA
$`31497_at`
[1] NA
$`31498_f_at`
[1] NA
$`31499_s_at`
[1] "O75015" "M9MML6" "A0A087WZR4" "A0A087WU90"
> names(sp_t) <- NULL
> for(i in 1:15) {
+ if(i == 9 || i == 10 || i ==12 ||
+ i ==13 || i == 14)
+ sp_t[[i]][1] <- " "
+ }
> sp
[[1]]
[1] "P21108"
[[2]]
[1] "Q10571"
[[3]]
[1] "Q9UHY8"
[[4]]
[1] "Q16444"
[[5]]
[1] " "
[[6]]
[1] "Q14524 " " Q8IZC9 " " Q8WTQ6 " " Q8WWN5 " " Q96J69"
[[7]]
[1] "Q14790"
[[8]]
[1] "Q9UBQ5"
[[9]]
[1] " "
[[10]]
[1] " "
[[11]]
[1] "P47992"
[[12]]
[1] " "
[[13]]
[1] "Q13065 " " Q8IYC5"
[[14]]
[1] "Q13070"
[[15]]
[1] "O75015"
> sp_t
[[1]]
[1] "P21108"
[[2]]
[1] "A0A024R1C3" "Q10571"
[[3]]
[1] "Q9UHY8"
[[4]]
[1] "P00558" "V9HWF4"
[[5]]
[1] "P54252" "A0A0A0MS38" "C9JQV6"
[[6]]
[1] "Q14524" "Q86V90" "H9KVD2" "E9PG18" "E9PHB6"
[6] "K4DIA1" "A0A0A0MT39"
[[7]]
[1] "A0A024R3Z8" "Q14790"
[[8]]
[1] "K7ERF1" "U3LUI4" "K7EK53" "Q9UBQ5"
[[9]]
[1] " "
[[10]]
[1] " "
[[11]]
[1] "Q9UBD3"
[[12]]
[1] " "
[[13]]
[1] " "
[[14]]
[1] " "
[[15]]
[1] "O75015" "M9MML6" "A0A087WZR4" "A0A087WU90"
> class(sp)
[1] "list"
> class(sp_t)
[1] "list"
SwissProt ID clear
> name_t <- as.character(unlist(lookUp(igenes, "hgu95av2", "GENENAME")))
> name_t
[1] "phosphoribosyl pyrophosphate synthetase 1 like 1"
[2] "MN1 proto-oncogene, transcriptional regulator"
[3] "fasciculation and elongation protein zeta 2"
[4] "phosphoglycerate kinase 1"
[5] "ataxin 3"
[6] "sodium voltage-gated channel alpha subunit 5"
[7] "caspase 8"
[8] "eukaryotic translation initiation factor 3 subunit K"
[9] NA
[10] NA
[11] "X-C motif chemokine ligand 2"
[12] NA
[13] NA
[14] NA
[15] "Fc fragment of IgG receptor IIIb"
> for(i in 1:length(names)) {
+ if(i == 9 || i == 10 || i ==12 ||
+ i ==13 || i == 14)
+ name_t[i] <- " "
+ }
> name
[1] "hypothetical protein LOC221823"
[2] "meningioma (disrupted in balanced translocation) 1"
[3] "fasciculation and elongation protein zeta 2 (zygin II)"
[4] "Phosphoglycerate kinase {alternatively spliced}"
[5] " "
[6] "sodium channel, voltage-gated, type V, alpha polypeptide"
[7] "caspase 8, apoptosis-related cysteine protease"
[8] "muscle specific gene"
[9] " "
[10] " "
[11] "chemokine (C motif) ligand 1"
[12] " "
[13] "G antigen 1"
[14] "G antigen 6"
[15] "Fc fragment of IgG, low affinity IIIb, receptor for (CD16)"
> name_t
[1] "phosphoribosyl pyrophosphate synthetase 1 like 1"
[2] "MN1 proto-oncogene, transcriptional regulator"
[3] "fasciculation and elongation protein zeta 2"
[4] "phosphoglycerate kinase 1"
[5] "ataxin 3"
[6] "sodium voltage-gated channel alpha subunit 5"
[7] "caspase 8"
[8] "eukaryotic translation initiation factor 3 subunit K"
[9] " "
[10] " "
[11] "X-C motif chemokine ligand 2"
[12] " "
[13] " "
[14] " "
[15] "Fc fragment of IgG receptor IIIb"
> class(name)
[1] "character"
> class(name_t)
[1] "character"
Gene Name clear
## 这些都是一毛一样的
dat <- exprs(sample.ExpressionSet)[igenes,1:10]
FC <- rowMeans(dat[igenes,1:5]) - rowMeans(dat[igenes,6:10])
pval <- esApply(sample.ExpressionSet[igenes,1:10], 1, function(x) t.test(x[1:5], x[6:10])$p.value)
tstat <- esApply(sample.ExpressionSet[igenes,1:10], 1, function(x) t.test(x[1:5], x[6:10])$statistic)
## 剩下的都对应地加上"_t"
genelist_t <- list(igenes, ug_t, ll_t, gb_t, sp_t)
filename_t <- "Interesting_genes_t.html"
title_t <- "An Artificial Set of Interesting Genes"
othernames_t <- list(name_t, round(tstat, 2), round(pval, 3), round(FC, 1), round(dat, 2))
head_t <- c("Probe ID", "UniGene", "LocusLink", "GenBank", "SwissProt", "Gene Name", "t-statistic", "p-value",
"Fold Change", "Sample 1", "Sample 2", "Sample 3", "Sample 4", "Sample 5", "Sample 6",
"Sample 7", "Sample 8", "Sample 9", "Sample 10")
repository_t <- list("affy", "ug", "en", "gb", "sp")
htmlpage(genelist_t, filename_t, title_t, othernames_t, head_t, repository = repository_t)
行吧。
最后,向大家隆重推荐生信技能树的一系列干货!
- 生信技能树全球公益巡讲:https://mp.weixin.qq.com/s/E9ykuIbc-2Ja9HOY0bn_6g
- B站公益74小时生信工程师教学视频合辑:https://mp.weixin.qq.com/s/IyFK7l_WBAiUgqQi8O7Hxw
- 招学徒:https://mp.weixin.qq.com/s/KgbilzXnFjbKKunuw7NVfw