Chemistry.AI | 基于图卷积神经网络(GCN)预测分子性质
GCN: Graph Convolutional Network(图卷积网络)
环境准备
- Python版本:Python 3.6.8
- PyTorch版本:PyTorch1.1.0
- RDKit版本:RDKit 2020.03.1
基于图卷积神经网络(GCN)预测分子性质
导入库
from rdkit import Chem
from rdkit.Chem.Crippen import MolLogP
import numpy as np
import torch
import time载入数据
max_natoms = 50
with open('smiles.txt') as f:
smiles = f.readlines()[:]
smiles = [s.strip() for s in smiles]
smiles = [s.split()[1] for s in smiles]
smiles = [s for s in smiles[:30000] if Chem.MolFromSmiles(s).GetNumAtoms()<50]
print ('Number of smiles:', len(smiles))计算分子指纹和描述符
Y = []
num_data = 20000
st = time.time()
for s in smiles[:num_data]:
m = Chem.MolFromSmiles(s)
logp = MolLogP(m)
Y.append(logp)
end = time.time()
print (f'Time:{(end-st):.3f}')
数据批处理
#Dataset
from torch.utils.data import Dataset, DataLoader
from rdkit.Chem.rdmolops import GetAdjacencyMatrix
class MolDataset(Dataset):
def __init__(self, smiles, properties, max_natoms, normalize_A=False):
self.smiles = smiles
self.properties = properties
self.max_natoms = max_natoms
def __len__(self):
return len(self.smiles)
def __getitem__(self, idx):
s = self.smiles[idx]
m = Chem.MolFromSmiles(s)
natoms = m.GetNumAtoms()
#adjacency matrix
A = GetAdjacencyMatrix(m) + np.eye(natoms)
A_padding = np.zeros((self.max_natoms, self.max_natoms))
A_padding[:natoms,:natoms] = A
#atom feature
X = [self.atom_feature(m,i) for i in range(natoms)]
for i in range(natoms, max_natoms):
X.append(np.zeros(28))
X = np.array(X)
sample = dict()
sample['X'] = torch.from_numpy(X)
sample['A'] = torch.from_numpy(A_padding)
sample['Y'] = self.properties[idx]
return sample
def normalize_A(A):
D = dfadfa(A)
A = D*DD
return A
def one_of_k_encoding(self, x, allowable_set):
if x not in allowable_set:
raise Exception("input {0} not in allowable set{1}:".format(x, allowable_set))
#print list((map(lambda s: x == s, allowable_set)))
return list(map(lambda s: x == s, allowable_set))
def one_of_k_encoding_unk(self, x, allowable_set):
"""Maps inputs not in the allowable set to the last element."""
if x not in allowable_set:
x = allowable_set[-1]
return list(map(lambda s: x == s, allowable_set))
def atom_feature(self, m, atom_i):
atom = m.GetAtomWithIdx(atom_i)
return np.array(self.one_of_k_encoding_unk(atom.GetSymbol(),
['C', 'N', 'O', 'S', 'F', 'P', 'Cl', 'Br', 'B', 'H']) +
self.one_of_k_encoding(atom.GetDegree(), [0, 1, 2, 3, 4, 5]) +
self.one_of_k_encoding_unk(atom.GetTotalNumHs(), [0, 1, 2, 3, 4]) +
self.one_of_k_encoding_unk(atom.GetImplicitValence(), [0, 1, 2, 3, 4, 5]) +
[atom.GetIsAromatic()]) # (10, 6, 5, 6, 1) --> total 28定义图卷积模型
#Model
import torch
import torch.nn as nn
import torch.nn.functional as F
class GConvRegressor(torch.nn.Module):
def __init__(self, n_feature=128, n_layer = 10):
super(GConvRegressor, self).__init__()
self.W = nn.ModuleList([nn.Linear(n_feature, n_feature) for _ in range(n_layer)])
self.embedding = nn.Linear(28, n_feature)
self.fc = nn.Linear(n_feature, 1)
def forward(self, x, A):
x = self.embedding(x)
for l in self.W:
x = l(x)
x = torch.einsum('ijk,ikl->ijl', (A.clone(), x))
x = F.relu(x)
x = x.mean(1)
retval = self.fc(x)
return retval训练模型
#Train model
import time
lr = 1e-4
model = GConvRegressor(128, 5).cuda()
#model initialize
for param in model.parameters():
if param.dim() == 1:
continue
nn.init.constant(param, 0)
else:
nn.init.xavier_normal_(param)
#Dataset
train_smiles = smiles[:19000]
test_smiles = smiles[19000:20000]
train_logp = Y[:19000]
test_logp = Y[19000:20000]
train_dataset = MolDataset(train_smiles, train_logp, max_natoms)
test_dataset = MolDataset(test_smiles, test_logp, max_natoms)
#Dataloader
train_dataloader = DataLoader(train_dataset, batch_size=128, num_workers=0)
test_dataloader = DataLoader(test_dataset, batch_size=128, num_workers=0)
optimizer = torch.optim.Adam(model.parameters(), lr=lr)
#optimizer = torch.optim.SGD(model.parameters(), lr=lr)
loss_fn = nn.MSELoss()
loss_list = []
st = time.time()
for epoch in range(20):
epoch_loss = []
for i_batch, batch in enumerate(train_dataloader):
x, y, A = \
batch['X'].cuda().float(), batch['Y'].cuda().float(), batch['A'].cuda().float()
pred = model(x, A).squeeze(-1)
loss = loss_fn(pred, y)
loss.backward()
torch.nn.utils.clip_grad_norm_(model.parameters(), 1.0)
optimizer.step()
loss_list.append(loss.data.cpu().numpy())
epoch_loss.append(loss.data.cpu().numpy())
if True: print (epoch, np.mean(np.array(epoch_loss)))
end = time.time()
print ('Time:', end-st)0 1.6517382
1 0.85889417
2 0.7216666
3 0.7328675
4 0.5567027
5 0.6174266
6 0.51587695
7 0.43320116
8 0.40305394
9 0.5059827
10 0.38442373
11 0.36464992
12 0.34420174
13 0.3333575
14 0.3868102
15 0.35386086
16 0.3727922
17 0.29447067
18 0.35121724
19 0.31530088
Time: 207.20838356018066
保存模型
#Save model
fn = 'save.pt'
torch.save(model.state_dict(), fn)加载模型
#Load model
model.load_state_dict(torch.load(fn))绘制损失曲线
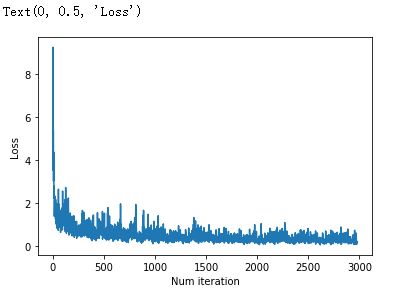
import matplotlib.pyplot as plt
plt.plot(loss_list)
plt.xlabel('Num iteration')
plt.ylabel('Loss')测试模型
#Test model
model.eval()
with torch.no_grad():
y_pred_train, y_pred_test = [], []
loss_train, loss_test = [], []
pred_train, pred_test = [], []
true_train, true_test = [], []
for batch in train_dataloader:
x, y, A = \
batch['X'].cuda().float(), batch['Y'].cuda().float(), batch['A'].cuda().float()
pred = model(x, A).squeeze(-1)
pred_train.append(pred.data.cpu().numpy())
true_train.append(y.data.cpu().numpy())
loss_train.append(loss_fn(y, pred).data.cpu().numpy())
for batch in test_dataloader:
x, y, A = \
batch['X'].cuda().float(), batch['Y'].cuda().float(), batch['A'].cuda().float()
pred = model(x, A).squeeze(-1)
pred_test.append(pred.data.cpu().numpy())
true_test.append(y.data.cpu().numpy())
loss_test.append(loss_fn(y, pred).data.cpu().numpy())
pred_train = np.concatenate(pred_train, -1)
pred_test = np.concatenate(pred_test, -1)
true_train = np.concatenate(true_train, -1)
true_test = np.concatenate(true_test, -1)
print ('Train loss:', np.mean(np.array(loss_train)))
print ('Test loss:', np.mean(np.array(loss_test)))Train loss: 0.23376586
Test loss: 0.24419878
plt.scatter(true_train, pred_train, s=1)
plt.scatter(true_test, pred_test, s=1)
plt.plot([-8,12], [-8,12])
plt.xlabel('True')
plt.ylabel('Pred')